1NX1
 
 | | Calpain Domain VI Complexed with Calpastatin Inhibitory Domain C (DIC) | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1NX3
 
 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1DOJ
 
 | | Crystal structure of human alpha-thrombin*RWJ-51438 complex at 1.7 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUGEN, ... | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Carson, M, DeLucas, L, Chattopadhyay, D. | | Deposit date: | 1999-12-21 | | Release date: | 2000-11-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human alpha-thrombin complexed with RWJ-51438 at 1.7 A: unusual perturbation of the 60A-60I insertion loop.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1NX0
 
 | | Structure of Calpain Domain 6 in Complex with Calpastatin DIC | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
5JX8
 
 | |
5JYE
 
 | |
5JX3
 
 | | Wild type D4 in orthorhombic space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2016-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
5JYA
 
 | |
5JX0
 
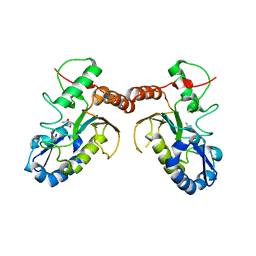 | | Temperature sensitive D4 mutant L110F | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
5JY6
 
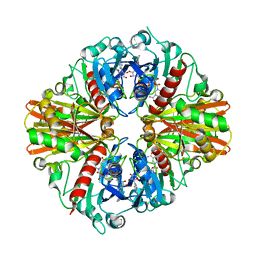 | |
5JYF
 
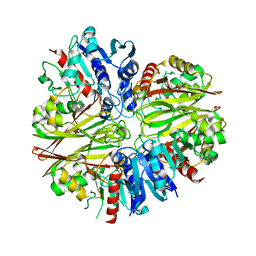 | |
4QCA
 
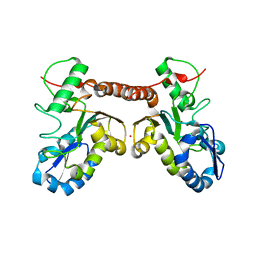 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant R167AD4 | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of three recombinant mutants of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3CL9
 
 | | Structure of bifunctional TcDHFR-TS in complex with MTX | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3CIF
 
 | | Crystal Structure of C153S mutant glyceraldehyde 3-phosphate dehydrogenase from Cryptosporidium parvum | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Cook, W.J, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An unexpected phosphate binding site in Glyceraldehyde 3-Phosphate Dehydrogenase: Crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme
BMC STRUCT.BIOL., 9, 2009
|
|
1DGM
 
 | | CRYSTAL STRUCTURE OF ADENOSINE KINASE FROM TOXOPLASMA GONDII | | Descriptor: | ACETIC ACID, ADENOSINE, ADENOSINE KINASE, ... | | Authors: | Cook, W.J, DeLucas, L.J, Chattopadhyay, D. | | Deposit date: | 1999-11-24 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenosine kinase from Toxoplasma gondii at 1.8 A resolution.
Protein Sci., 9, 2000
|
|
1EOU
 
 | | CRYSTAL STRUCTURE OF HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH AN ANTICONVULSANT SUGAR SULFAMATE | | Descriptor: | CARBONIC ANHYDRASE II (CA II), SULFAMIC ACID 2,3-O-(1-METHYLETHYLIDENE)-4,5-O-SULFONYL-BETA-FRUCTOPYRANOSE ESTER, ZINC ION | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Chattopadhyay, D. | | Deposit date: | 2000-03-24 | | Release date: | 2002-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human carbonic anhydrase II complexed with an anti-convulsant sugar sulphamate.
Biochem.J., 361, 2002
|
|
1NC6
 
 | | Potent, small molecule inhibitors of human mast cell tryptase. Anti-asthmatic action of a dipeptide-based transition state analogue containing benzothiazole ketone | | Descriptor: | (2S,4R)-1-ACETYL-N-[(1S)-4-[(AMINOIMINOMETHYL)AMINO]-1-(2-BENZOTHIAZOLYLCARBONYL)BUTYL]-4-HYDROXY-2-PYRROLIDINECARBOXAMIDE, CALCIUM ION, SULFATE ION, ... | | Authors: | Costanzo, M.J, Yabut, S.C, Almond Jr, H.R, Andrade-Gordon, P, Corcoran, T.W, de Garavilla, L, Kauffman, J.A, Abraham, W.M, Recacha, R, Chattopadhyay, D, Maryanoff, B.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, Small-Molecule Inhibitors of Human Mast Cell Tryptase. Antiasthmatic Action of a Dipeptide-Based Transition-State Analogue Containing a Benzothiazole Ketone.
J.Med.Chem., 46, 2003
|
|
1KYN
 
 | | Cathepsin-G | | Descriptor: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
2OKE
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OKD
 
 | |
2OKB
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OL1
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OL0
 
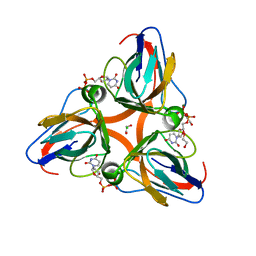 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DEOXYURIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OWQ
 
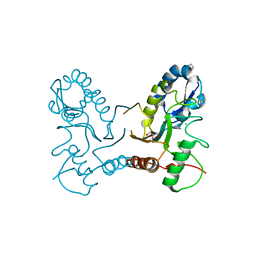 | |
