3LTD
 
 | | X-ray structure of a non-biological ATP binding protein determined at 2.8 A by multi-wavelength anomalous dispersion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LTC
 
 | | X-ray structure of a non-biological ATP binding protein determined in the presence of 10 mM ATP at 2.0 A by multi-wavelength anomalous dispersion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LTA
 
 | | Crystal structure of a non-biological ATP binding protein with a TYR-PHE mutation within the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
6CIO
 
 | | Pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica with lactyl-TPP bound | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2018-02-24 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase fromMoorella thermoacetica.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3M1D
 
 | | Structure of BIR1 from cIAP1 | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Mace, P.D, Day, C.L. | | Deposit date: | 2010-03-04 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric recruitment of cIAPs by TRAF2
J.Mol.Biol., 400, 2010
|
|
4OEY
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OH6
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, Protein BUD31 homolog, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OKB
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, Protein BUD31 homolog, R-BICALUTAMIDE, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OOE
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203Y mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4PKF
 
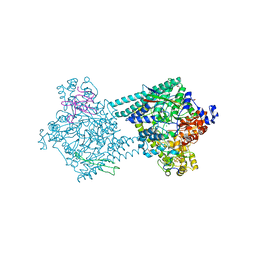 | | Benzylsuccinate synthase alpha-beta-gamma complex | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PKY
 
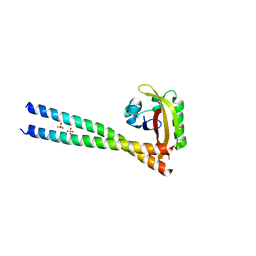 | | ARNT/HIF transcription factor/coactivator complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, SULFATE ION, ... | | Authors: | Tomchick, D.R, Partch, C.L, Gardner, K.H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coiled-coil Coactivators Play a Structural Role Mediating Interactions in Hypoxia-inducible Factor Heterodimerization.
J.Biol.Chem., 290, 2015
|
|
6VWZ
 
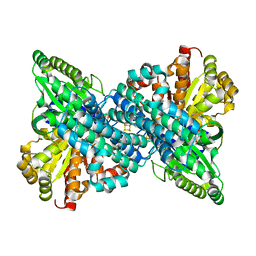 | |
6VXE
 
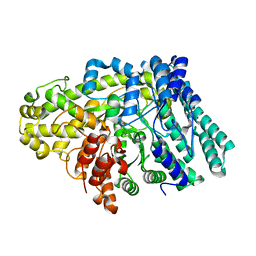 | |
6VXC
 
 | |
6W4X
 
 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C.L. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
6W69
 
 | | The structure of F64, S172A Keap1-BTB domain | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6W68
 
 | |
6W9D
 
 | |
6VUE
 
 | | wild-type choline TMA lyase in complex with 1-methyl-1,2,3,6-tetrahydropyridin-3-ol | | Descriptor: | (3S)-1-methyl-1,2,3,6-tetrahydropyridin-3-ol, Choline trimethylamine-lyase, SODIUM ION | | Authors: | Ortega, M.A, Drennan, C.L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
Acs Med.Chem.Lett., 11, 2020
|
|
1NPB
 
 | | Crystal structure of the fosfomycin resistance protein from transposon Tn2921 | | Descriptor: | GLYCEROL, SULFATE ION, fosfomycin-resistance protein | | Authors: | Pakhomova, S, Rife, C.L, Armstrong, R.N, Newcomer, M.E. | | Deposit date: | 2003-01-17 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of fosfomycin resistance protein FosA from transposon Tn2921.
Protein Sci., 13, 2004
|
|
1NM6
 
 | | thrombin in complex with selective macrocyclic inhibitor at 1.8A | | Descriptor: | (11S)-11-BENZYL-6-CHLORO-1,2,10,11,12,13,14,15,16,17,18,19-DODECAHYDRO-5,9-METHANO-2,5,8,10,13,17-BENZOHEXAAZACYCLOHENICOSINE-3,24-DIONE, Hirudin, thrombin | | Authors: | Nantermet, P.G, Barrow, J.C, Newton, C.L, Pellicore, J.M, Young, M, Lewis, S.D, Lucas, B.J, Krueger, J.A, McMasters, D.R, Yan, Y, Kuo, L.C, Vacca, J.P, Selnick, H.G. | | Deposit date: | 2003-01-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of potent and selective macrocyclic thrombin inhibitors
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NZ2
 
 | | K45E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, M, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NT1
 
 | | thrombin in complex with selective macrocyclic inhibitor | | Descriptor: | (6R,21AS)-17-CHLORO-6-CYCLOHEXYL-2,3,6,7,10,11,19,20-OCTAHYDRO-1H,5H-PYRROLO[1,2-K][1,4,8,11,14]BENZOXATETRAAZA-CYCLOHEPTADECINE-5,8,12,21(9H,13H,21AH)-TETRONE, Hirudin, thrombin | | Authors: | Nantermet, P.G, Barrow, J.C, Newton, C.L, Pellicore, J.M, Young, M, Lewis, S.D, Lucas, B.J, Krueger, J.A, McMasters, D.R, Yan, Y, Kuo, L.C, Vacca, J.P, Selnick, H.G. | | Deposit date: | 2003-01-28 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent and selective macrocyclic thrombin inhibitors
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NZ5
 
 | | The Horse heart myoglobin variant K45E/K63E complexed with Manganese | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1O1R
 
 | | Structure of FPT bound to GGPP | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Protein farnesyltransferase alpha subunit, Protein farnesyltransferase beta subunit, ... | | Authors: | Turek-Etienne, T.C, Strickland, C.L, Distefano, M.D. | | Deposit date: | 2003-01-14 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Studies with Prenyl Diphosphate Analogues Provide Insights
into Isoprenoid Recognition by Protein Farnesyl Transferase
Biochemistry, 42, 2003
|
|
