7U96
 
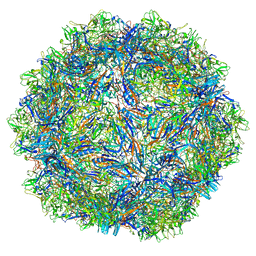 | | SAAV pH 5.5 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U94
 
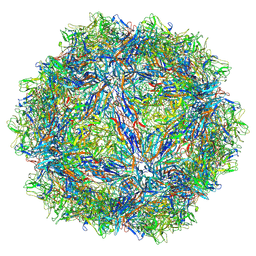 | | SAAV pH 7.4 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7AA9
 
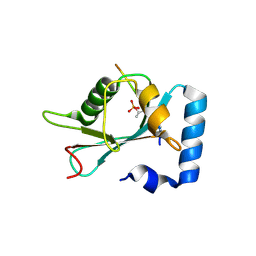 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA8
 
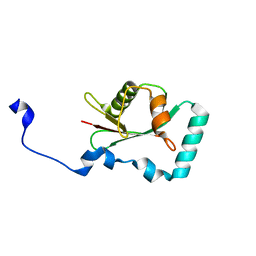 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
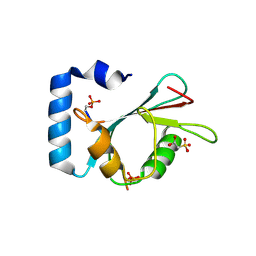 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7ABM
 
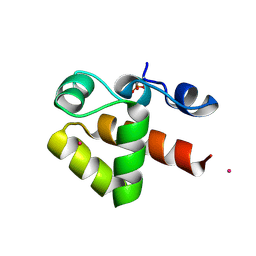 | | X-ray structure of phosphorylated Barrier-to-autointegration factor (BAF) | | Descriptor: | Barrier-to-autointegration factor, CESIUM ION | | Authors: | Zinn-Justin, S, Marcelot, A, Le Du, M.H, Ropars, V. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
280D
 
 | |
7ACZ
 
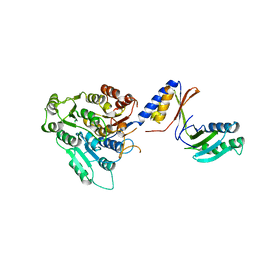 | |
7ACY
 
 | |
7ACX
 
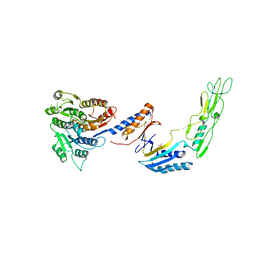 | | H/L (SLPH/SLPL) complex from C. difficile (R7404 strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-layer protein, SULFATE ION | | Authors: | Lanzoni-Mangutchi, P, Barwinska-Sendra, A, Basle, A, El Omari, K, Wagner, A, Salgado, P.S. | | Deposit date: | 2020-09-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and assembly of the S-layer in C. difficile.
Nat Commun, 13, 2022
|
|
7ACV
 
 | |
7ACW
 
 | |
5KX9
 
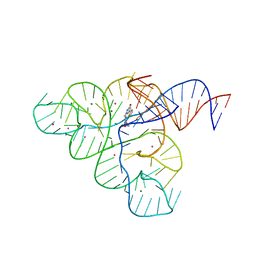 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6P)-2-{(3S)-1-[(2-methoxypyrimidin-5-yl)methyl]piperidin-3-yl}-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomic resolution mechanistic studies of ribocil: A highly selective unnatural ligand mimic of the E. coli FMN riboswitch.
Rna Biol., 13, 2016
|
|
7U99
 
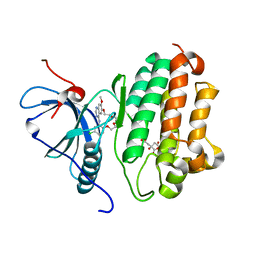 | | EGFR kinase in complex with a macrocyclic inhibitor | | Descriptor: | 19-chloro-22-methoxy-8,9,11,12,14,15-hexahydro-21H-4,6-ethenopyrimido[5,4-m][1,4,7,10,15]benzotetraoxazacycloheptadecine, CITRATE ANION, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclization of Quinazoline-Based EGFR Inhibitors Leads to Exclusive Mutant Selectivity for EGFR L858R and Del19.
J.Med.Chem., 65, 2022
|
|
7U98
 
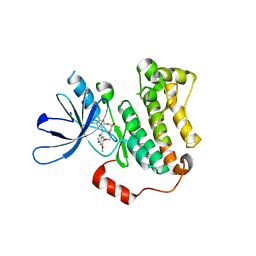 | | EGFR(T790M/V948R) in complex with a macrocyclic inhibitor | | Descriptor: | 19-chloro-18-fluoro-22-methoxy-8,9,11,12,14,15-hexahydro-21H-4,6-ethenopyrimido[5,4-m][1,4,7,10,15]benzotetraoxazacycloheptadecine, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Macrocyclization of Quinazoline-Based EGFR Inhibitors Leads to Exclusive Mutant Selectivity for EGFR L858R and Del19.
J.Med.Chem., 65, 2022
|
|
7U49
 
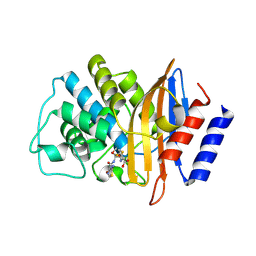 | | DFC-CTX-M-15 | | Descriptor: | (2R,4S,5R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-oxoethyl]-5-(sulfanylmethyl)-1,3-thiazinane-4-carboxylic acid, Beta-lactamase | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U48
 
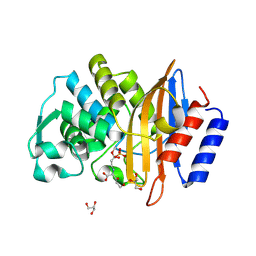 | | Clavulanic acid-CTX-M-15 | | Descriptor: | (E)-5-hydroxy-3-oxo-N-(3-oxopropylidene)-L-norvaline, Beta-lactamase, GLYCEROL, ... | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U0K
 
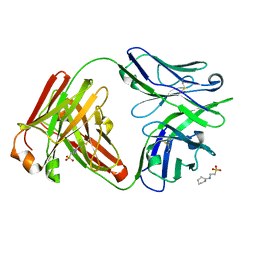 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
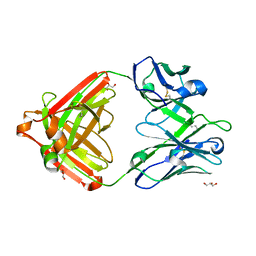 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7VX2
 
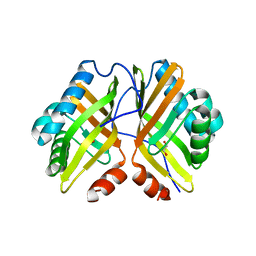 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWD
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWM
 
 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-11 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
