8ASM
 
 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8BA6
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
8BAJ
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
8BSG
 
 | | COMPLEX OF LEPORINE SERUM ALBUMIN WITH DICLOFENAC | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J, Zielinski, K. | | Deposit date: | 2022-11-25 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
8BR9
 
 | |
8BJ7
 
 | |
8BJ8
 
 | |
8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8BBY
 
 | |
2I3H
 
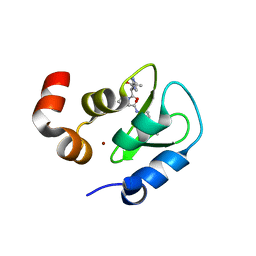 | | Structure of an ML-IAP/XIAP chimera bound to a 4-mer peptide (AVPW) | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AVPW peptide, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
2I3I
 
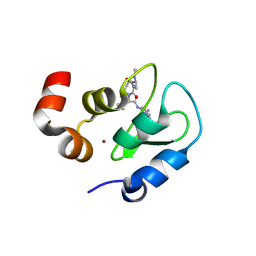 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | (3R,6R,9AR)-2,2-DIMETHYL-6-[(N-METHYL-L-ALANYL)AMINO]-N-(3-METHYL-1-PHENYL-1H-PYRAZOL-5-YL)-5-OXO-2,3,5,6,9,9A-HEXAHYDRO[1,3]THIAZOLO[3,2-A]AZEPINE-3-CARBOXAMIDE, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
8BAG
 
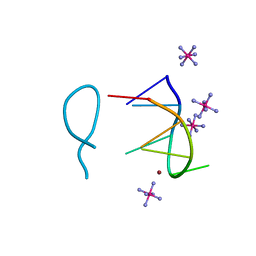 | |
8BAE
 
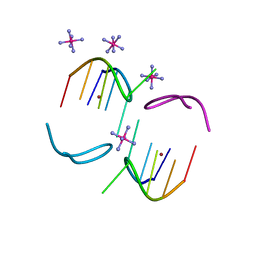 | |
8B7P
 
 | | Crystal structure of an AA9 LPMO from Aspergillus nidulans, AnLPMOC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Endo-beta-1,4-glucanase D | | Authors: | Males, A, Rafael Fanchini Terrasan, C, Davies, G.J, Walton, P.H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterisation of lytic polysaccharide monooxygenases from Aspergillus nidulans
To Be Published
|
|
8BAF
 
 | |
2HH5
 
 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8C9J
 
 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
8C3N
 
 | |
8COM
 
 | | Structure of the Nucleosome Core Particle from Trypanosoma brucei | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Burdett, H, Deak, G, Wilson, M.D. | | Deposit date: | 2023-02-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Histone divergence in trypanosomes results in unique alterations to nucleosome structure.
Nucleic Acids Res., 51, 2023
|
|
8CPH
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8AH5
 
 | |
8AH6
 
 | |
8AH7
 
 | |
8AH4
 
 | |
8AH8
 
 | |
