1L22
 
 | |
1L23
 
 | |
3G0W
 
 | |
2VKI
 
 | | Structure of the PDK1 PH domain K465E mutant | | Descriptor: | 3-PHOPSHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Bayascas, J.R, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutation of the Pdk1 Ph Domain Inhibits Protein Kinase B/Akt, Leading to Small Size and Insulin Resistance.
Mol.Cell.Biol., 28, 2008
|
|
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
1L20
 
 | |
3GTA
 
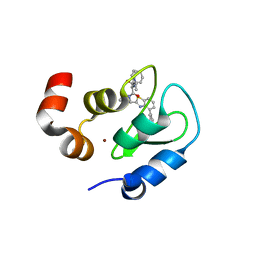 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3GT9
 
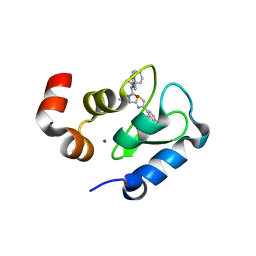 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | Baculoviral IAP repeat-containing 7, N-{(1S)-1-cyclohexyl-2-[(2S)-2-(4-naphthalen-1-yl-1,3-thiazol-2-yl)pyrrolidin-1-yl]-2-oxoethyl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3H5Z
 
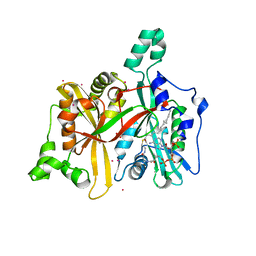 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
6ITK
 
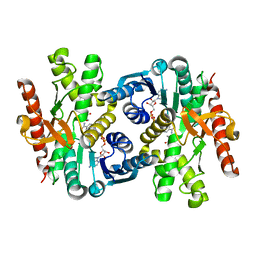 | |
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
6J4R
 
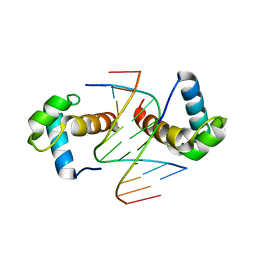 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J6Y
 
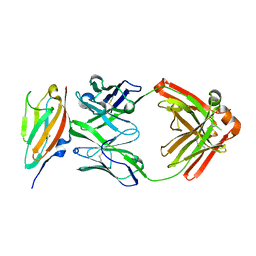 | | FGFR4 D2 - Fab complex | | Descriptor: | Fab Heavy chain, Fab light chain, Fibroblast growth factor receptor 4 | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Preclinical Development of U3-1784, a Novel FGFR4 Antibody Against Cancer, and Avoidance of Its On-target Toxicity.
Mol.Cancer Ther., 18, 2019
|
|
6J4K
 
 | |
6J5B
 
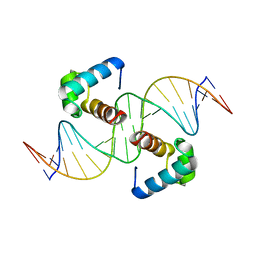 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
1GZ8
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 2-Amino-6-(3'-methyl-2'-oxo)butoxypurine | | Descriptor: | 1-[(2-AMINO-6,9-DIHYDRO-1H-PURIN-6-YL)OXY]-3-METHYL-2-BUTANOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davies, T, Endicott, J, Johnson, L, Noble, M, Tucker, J. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1ULA
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6IBP
 
 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6MBJ
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MDZ
 
 | |
6M8Q
 
 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
6MFL
 
 | | Structure of siderophore binding protein BauB bound to a complex between two molecules of acinetobactin and ferric iron. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Siderophore Binding Protein BauB, ... | | Authors: | Gulick, A.M, Bailey, D.C, Wencewicz, T.A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Siderophore Binding Protein BauB Bound to an Unusual 2:1 Complex Between Acinetobactin and Ferric Iron.
Biochemistry, 57, 2018
|
|
