4XNW
 
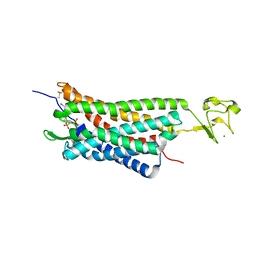 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
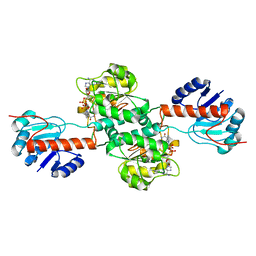
4XKJ
 
 | |
8J45
 
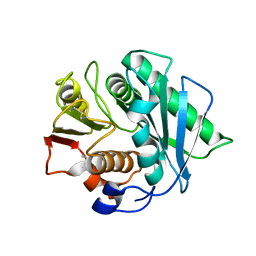 | | Crystal structure of a Pichia pastoris-expressed IsPETase variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, He, H.L, Long, X, Niu, D, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chem Eng J, 2023
|
|
7QTY
 
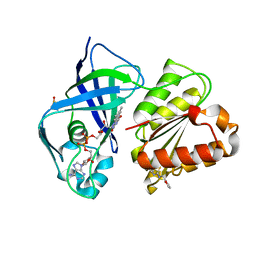 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | 1-(furan-2-ylmethyl)-3-(2-methylphenyl)thiourea, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU5
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4XR8
 
 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
7QLA
 
 | | Structure of the Rab GEF complex Mon1-Ccz1 | | Descriptor: | Ccz1, Vacuolar fusion protein MON1 | | Authors: | Klink, B.U, Herrmann, E, Antoni, C, Langemeyer, L, Kiontke, S, Gatsogiannis, C, Ungermann, C, Raunser, S, Kuemmel, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QU3
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU0
 
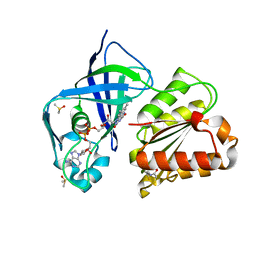 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2X34
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | CELLULOSE-BINDING PROTEIN, X158, Ubiquinone-8 | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
4XE5
 
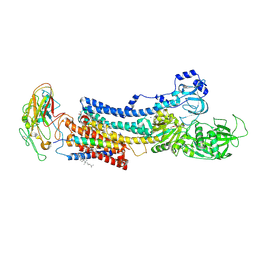 | | Crystal structure of the Na,K-ATPase from bovine | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, OUABAIN, ... | | Authors: | Gregersen, J.L, Mattle, D, Fedosova, N.U, Nissen, P, Reinhard, L. | | Deposit date: | 2014-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | Isolation, crystallization and crystal structure determination of bovine kidney Na(+),K(+)-ATPase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4XIN
 
 | |
7QEP
 
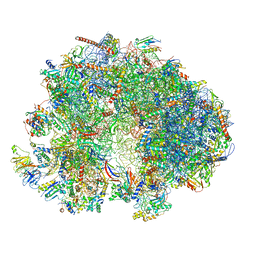 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
4XRA
 
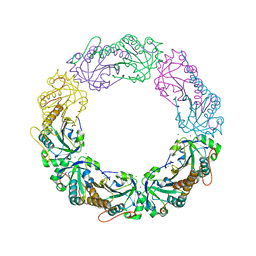 | | Salmonella typhimurium AhpC T43S mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Perkins, A, Brereton, A.E, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4XBI
 
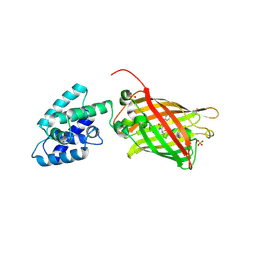 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
8JE0
 
 | | A novel amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, Amidase, ZINC ION | | Authors: | Ma, D, Feng, R. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel amidohydrolase catalyze the degradation of PAM by Klebsiella sp. PCX
To Be Published
|
|
4PTI
 
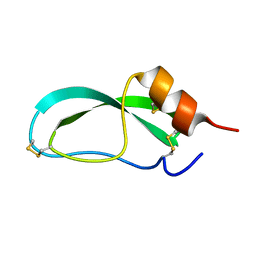 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
4XRD
 
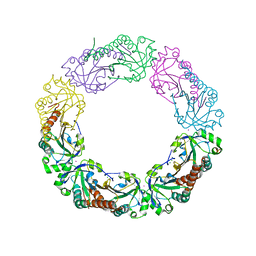 | | Salmonella typhimurium AhpC W169F mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4XS4
 
 | | Salmonella typhimurium AhpC C165S mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, POTASSIUM ION | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4XTS
 
 | | Salmonella typhimurium AhpC T43A mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4XS6
 
 | | Salmonella typhimurium AhpC W81F mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, POTASSIUM ION | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4XT3
 
 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
4XXF
 
 | | L-fuculose 1-phosphate aldolase from Glaciozyma antarctica PI12 | | Descriptor: | Fuculose-1-phosphate aldolase, ZINC ION | | Authors: | Jaafar, N.R, Abu Bakar, F.D, Abdul Murad, A.M, Illias, R, Littler, D, Beddoe, T, Rossjohn, J, Mahadi, M.N. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of fuculose aldolase from the Antarctic psychrophilic yeast Glaciozyma antarctica PI12.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8IZZ
 
 | |
2UX8
 
 | | Crystal Structure of Sphingomonas elodea ATCC 31461 Glucose-1- phosphate uridylyltransferase in Complex with glucose-1-phosphate. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Aragao, D, Fialho, A.M, Marques, A.R, Frazao, C, Sa-Correia, I, Mitchell, E.P. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Complex of Sphingomonas Elodea Atcc 31461 Glucose-1-Phosphate Uridylyltransferase with Glucose-1-Phosphate Reveals a Novel Quaternary Structure, Unique Among Nucleoside Diphosphate-Sugar Pyrophosphorylase Members.
J.Bacteriol., 189, 2007
|
|
