6MMZ
 
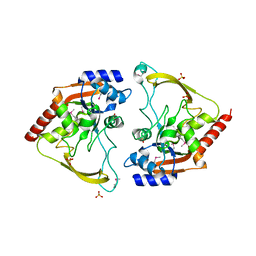 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN4
 
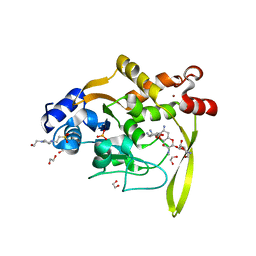 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with apramycin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APRAMYCIN, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
4IMY
 
 | |
3RQ9
 
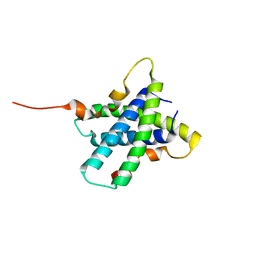 | | Structure of Tsi2, a Tse2-immunity protein from Pseudomonas aeruginosa | | Descriptor: | Type VI secretion immunity protein | | Authors: | Li, M, Le Trong, I, Stenkamp, R.E, Mougous, J.D. | | Deposit date: | 2011-04-27 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis for Type VI Secretion Effector Recognition by a Cognate Immunity Protein.
Plos Pathog., 8, 2012
|
|
6G74
 
 | |
6F48
 
 | | Structure of quinolinate synthase with reaction intermediates X and Y | | Descriptor: | 2-imino,3-carboxy,5-hydroxy,6-oxo hexanoic acid, 5-hydroxy,-4,5-dihydroquinolinate, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Trapping of Reaction Intermediates in Quinolinic Acid Synthesis by NadA.
ACS Chem. Biol., 13, 2018
|
|
6F4L
 
 | |
6F4D
 
 | |
7B52
 
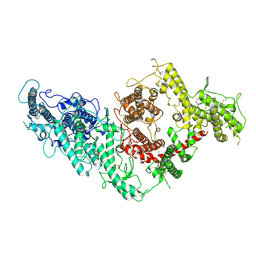 | | VAR2CSA full ectodomain | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Gourdon, P.E, Dagil, R, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
7B54
 
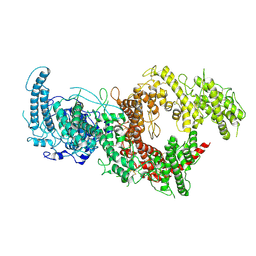 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9CPJ
 
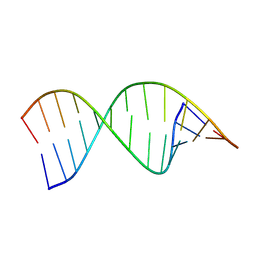 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPG
 
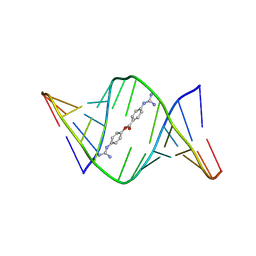 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPD
 
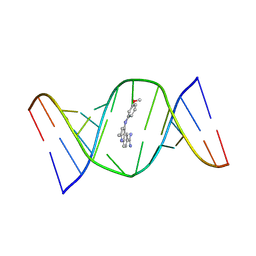 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
5LQM
 
 | |
5LQS
 
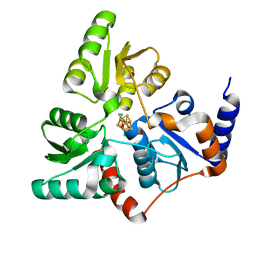 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
7L7F
 
 | |
7L7K
 
 | |
5F35
 
 | | Structure of quinolinate synthase in complex with citrate | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F3D
 
 | | Structure of quinolinate synthase in complex with reaction intermediate W | | Descriptor: | 2-IMINO,3-CARBOXY,5-OXO,6-HYDROXY HEXANOIC ACID, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F33
 
 | | Structure of quinolinate synthase in complex with phosphoglycolohydroxamate | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHOGLYCOLOHYDROXAMIC ACID, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
4D8D
 
 | |
1AVV
 
 | | HIV-1 NEF PROTEIN, UNLIGANDED CORE DOMAIN | | Descriptor: | NEGATIVE FACTOR | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1997-09-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling.
Structure, 5, 1997
|
|
1AVZ
 
 | |
