5CI1
 
 | | Ribonucleotide reductase Y122 2,3-F2Y variant | | Descriptor: | CHLORIDE ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI0
 
 | | Ribonucleotide reductase Y122 3,5-F2Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5TQN
 
 | | Lipoxygenase-1 (soybean) L546A mutant at 293K | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
5UFV
 
 | |
5TQO
 
 | | Lipoxygenase-1 (soybean) L546A/L754A mutant at 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
5TR0
 
 | | Lipoxygenase-1 (soybean) L754A mutant at 293K | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
6WZ3
 
 | | Cu-bound structure of the engineered protein trimer, TriCyt3 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6WZA
 
 | | Ni-bound structure of an engineered metal-dependent protein trimer, TriCyt1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, HEME C, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6X0Q
 
 | |
6WZ2
 
 | |
6WZ7
 
 | | Mn-bound structure of a TriCyt3 variant | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6WZC
 
 | |
6X0R
 
 | |
6WZ0
 
 | |
6WZ1
 
 | |
6WYU
 
 | | Crystallographic trimer of metal-free TriCyt2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HEME C, SULFATE ION, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6X7E
 
 | | Co-bound structure of an engineered protein trimer, TriCyt3, with delta isomerism at the hexahistidine coordination site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9987 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8SM6
 
 | | Aerobic, Diiron(III)-metalated SfbO | | Descriptor: | Amidohydrolase family protein, FE (III) ION, SULFATE ION | | Authors: | Liu, C, Powell, M.M, Rittle, J. | | Deposit date: | 2023-04-25 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Bioinformatic Discovery of a Cambialistic Monooxygenase.
J.Am.Chem.Soc., 146, 2024
|
|
8SM8
 
 | |
8SMA
 
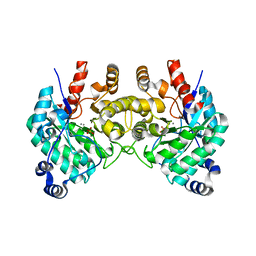 | | SfbO with di-Manganese cofactor | | Descriptor: | Amidohydrolase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Liu, C, Rittle, J. | | Deposit date: | 2023-04-25 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bioinformatic Discovery of a Cambialistic Monooxygenase.
J.Am.Chem.Soc., 146, 2024
|
|
8SM9
 
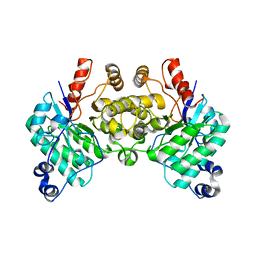 | |
8SM7
 
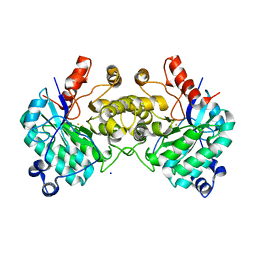 | |
