3M5V
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-28 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
4JRO
 
 | | Crystal structure of 3-oxoacyl-[acyl-carrier protein]reductase (FabG)from Listeria monocytogenes in complex with NADP+ | | Descriptor: | FabG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Zheng, H, Cooper, D.R, Osinski, T, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of 3-oxoacyl-[acyl-carrier protein]reductase (FabG)from Listeria monocytogenes in complex with NADP+
To be Published
|
|
3OO2
 
 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | Descriptor: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3KB8
 
 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
3N0L
 
 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
7MQ5
 
 | | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14 | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14
To Be Published
|
|
3GIU
 
 | | 1.25 Angstrom Crystal Structure of Pyrrolidone-Carboxylate Peptidase (pcp) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Pyrrolidone-Carboxylate Peptidase (pcp) from Staphylococcus aureus
TO BE PUBLISHED
|
|
3OWA
 
 | | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis | | Descriptor: | Acyl-CoA dehydrogenase, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis
To be Published
|
|
3N5M
 
 | | Crystals structure of a Bacillus anthracis aminotransferase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, DiLeo, R, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystals structure of a Bacillus anthracis aminotransferase
TO BE PUBLISHED
|
|
3K96
 
 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
3PEI
 
 | | Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Cytosol Aminopeptidase from
Francisella tularensis
To be Published
|
|
7M5F
 
 | | Contact-dependent inhibition system from Serratia marcescens BWH57 | | Descriptor: | CdiI, MALONATE ION, Toxin CdiA | | Authors: | Michalska, K, Nutt, W, Stols, L, Jedrzejczak, R, Hayes, C.S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Contact-dependent inhibition system from Serratia marcescens
To Be Published
|
|
3O2R
 
 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
3H5Q
 
 | | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SULFATE ION, THYMIDINE | | Authors: | Shumilin, I.A, Zimmerman, M, Cymborowski, M, Skarina, T, Onopriyenko, O, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3OJC
 
 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3OQ3
 
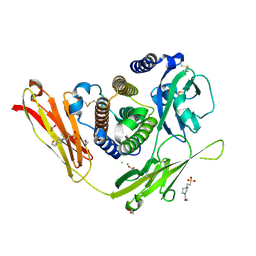 | |
3KY7
 
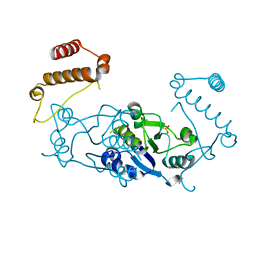 | | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252 | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, See, R, Zoraghi, R, Reiner, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252
To be Published
|
|
3HVU
 
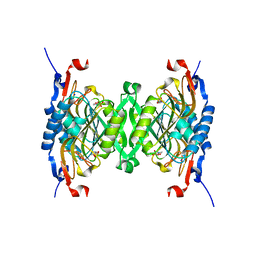 | | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypoxanthine phosphoribosyltransferase, SODIUM ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES)
TO BE PUBLISHED
|
|
3IST
 
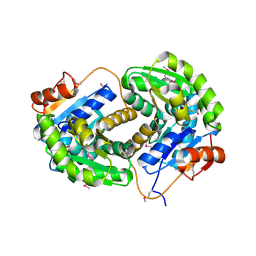 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | Descriptor: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
4FK1
 
 | | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis
To be Published
|
|
3O7M
 
 | | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor'
TO BE PUBLISHED
|
|
4IJK
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), SODIUM ION | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I.G, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695
To be Published
|
|
