8R08
 
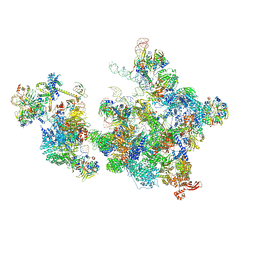 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPK
 
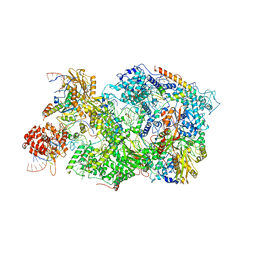 | | Cryo-EM Structure of Pre-B+5'ss Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8SXS
 
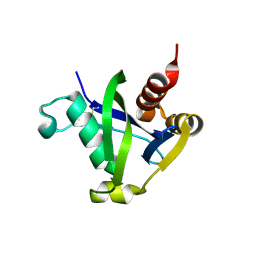 | |
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
1IW2
 
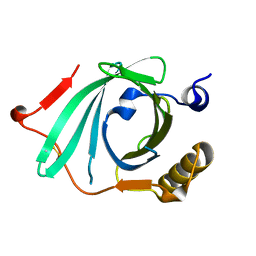 | | X-ray structure of Human Complement Protein C8gamma at pH=7.O | | Descriptor: | Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
4XC7
 
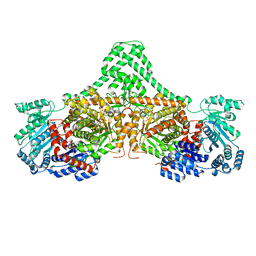 | |
4XYN
 
 | | X-ray structure of Ca(2+)-S100B with human RAGE-derived W61 peptide | | Descriptor: | CALCIUM ION, Protein S100-B, Receptor for advanced glycation endproducts-derived peptide (W61) | | Authors: | Jensen, J.L, Indurthi, V.S.K, Neau, D, Vetter, S.W, Colbert, C.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the binding of the human receptor for advanced glycation end products (RAGE) by S100B, as revealed by an S100B-RAGE-derived peptide complex.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJK
 
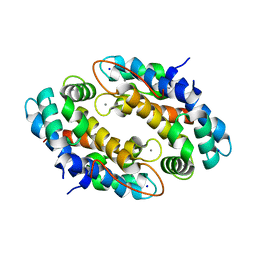 | | Crystal structure of Mn(II) Ca(II) Na(I) bound calprotectin | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Protein S100-A8, ... | | Authors: | Drennan, C.L, Bowman, S.E.J. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Manganese Binding Properties of Human Calprotectin under Conditions of High and Low Calcium: X-ray Crystallographic and Advanced Electron Paramagnetic Resonance Spectroscopic Analysis.
J.Am.Chem.Soc., 137, 2015
|
|
4XC8
 
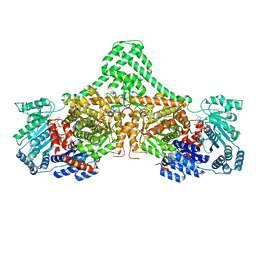 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XHM
 
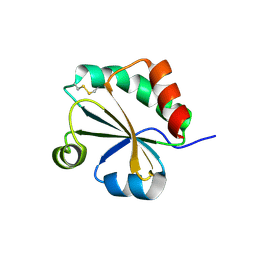 | |
4XC6
 
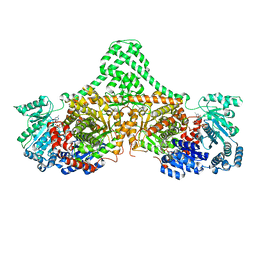 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZC7
 
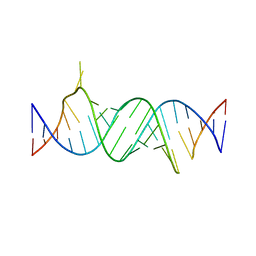 | | Paromomycin bound to a leishmanial ribosomal A-site | | Descriptor: | PAROMOMYCIN, RNA duplex | | Authors: | Shalev, M, Rozenberg, H, Jaffe, C.L, Adir, N, Baasov, T. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural basis for selective targeting of leishmanial ribosomes: aminoglycoside derivatives as promising therapeutics.
Nucleic Acids Res., 43, 2015
|
|
4ZWI
 
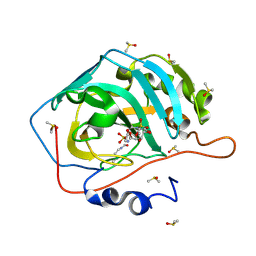 | | Surface Lysine Acetylated Human Carbonic Anhydrase II in Complex with a Sulfamate-Based Inhibitor | | Descriptor: | (6R)-1-O-acetyl-2,6-anhydro-6-{[4-(sulfamoyloxy)piperidin-1-yl]sulfonyl}-L-glucitol, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, M. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Observed surface lysine acetylation of human carbonic anhydrase II expressed in Escherichia coli.
Protein Sci., 24, 2015
|
|
4ZWZ
 
 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX Using Structure-Activity Relationships of Glucosyl-Based Sulfamates.
J.Med.Chem., 58, 2015
|
|
2VJQ
 
 | | Formyl-CoA transferase mutant variant W48Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMYL-COENZYME A TRANSFERASE | | Authors: | Toyota, C.G, Berthold, C.L, Gruez, A, Jonsson, S, Lindqvist, Y, Cambillau, C, Richards, N.G.J. | | Deposit date: | 2007-12-11 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Substrate Specificity and Kinetic Behavior of Escherichia Coli Yfdw and Oxalobacter Formigenes Formyl Coenzyme a Transferase.
J.Bacteriol., 190, 2008
|
|
2VJM
 
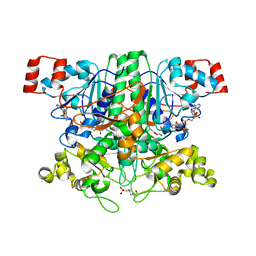 | | Formyl-CoA transferase with aspartyl-formyl anhydide intermediate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
5E1V
 
 | |
2VOI
 
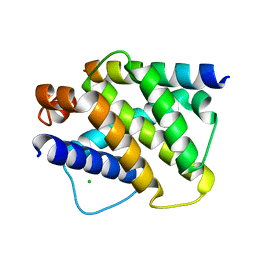 | | Structure of mouse A1 bound to the Bid BH3-domain | | Descriptor: | BCL-2-RELATED PROTEIN A1, BH3-INTERACTING DOMAIN DEATH AGONIST P13, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VO2
 
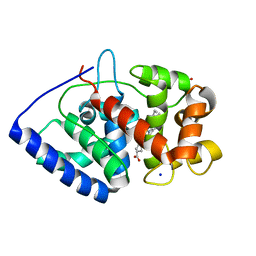 | | Crystal structure of soybean ascorbate peroxidase mutant W41A subjected to low dose X-rays | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
6DBD
 
 | | Crystal Structure of VHH R326 | | Descriptor: | ACETATE ION, SODIUM ION, nanobody VHH R326 | | Authors: | Brooks, C.L, Toride King, M, Huh, I. | | Deposit date: | 2018-05-03 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of VHH-mediated neutralization of the food-borne pathogenListeria monocytogenes.
J. Biol. Chem., 293, 2018
|
|
6DDK
 
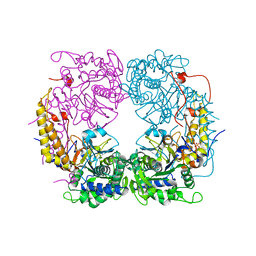 | | Crystal structure of the double mutant (D52N/R367Q) of the full-length NT5C2 in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE1
 
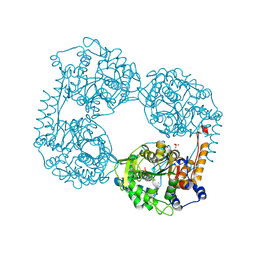 | | Crystal structure of the single mutant (D52N) of the full-length NT5C2 in the active state | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDH
 
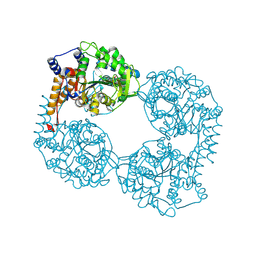 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, INOSINIC ACID | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE0
 
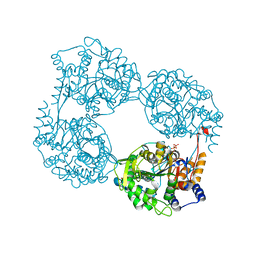 | | Crystal structure of the single mutant (D52N) of NT5C2-Q523X in the active state | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
