1M9R
 
 | | human endothelial nitric oxide synthase with 3-Bromo-7-Nitroindazole bound | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1MDO
 
 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1VVJ
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.440001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HQ1
 
 | | STRUCTURAL AND ENERGETIC ANALYSIS OF RNA RECOGNITION BY A UNIVERSALLY CONSERVED PROTEIN FROM THE SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Sagar, M.B, Doudna, J.A. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
J.Mol.Biol., 307, 2001
|
|
1TKK
 
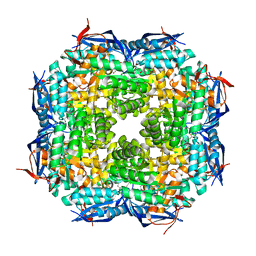 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
3NQD
 
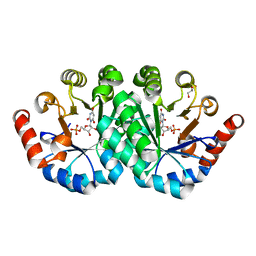 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
1GO2
 
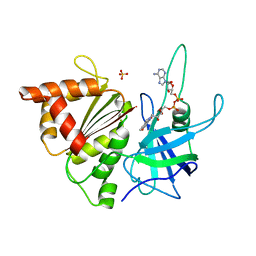 | | Structure of Ferredoxin-NADP+ Reductase with Lys 72 replaced by Glu (K72E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-10-15 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners
Proteins: Struct.,Funct., Genet., 59, 2005
|
|
1SSZ
 
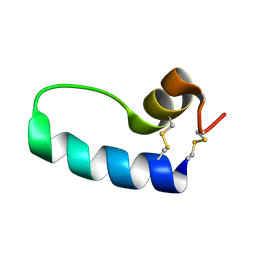 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2019-04-24 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
1GR1
 
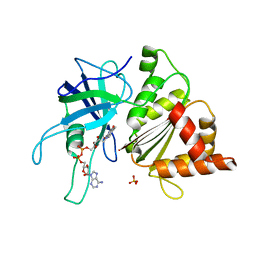 | | Structure of Ferredoxin-NADP+ Reductase with Glu 139 replaced by Lys (E139K) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Role of Glutamic Acid 139 of Anabena Ferrodoxin-Nadp+ Reductase in the Interaction with Substrates
Eur.J.Biochem., 269, 2002
|
|
1GRP
 
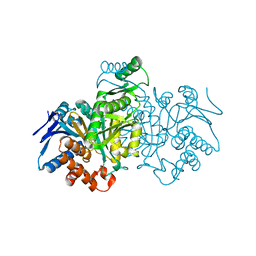 | |
1SZU
 
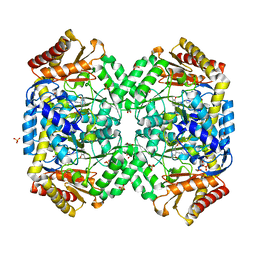 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1T3G
 
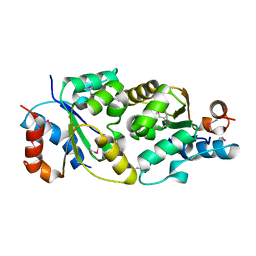 | |
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1T6G
 
 | | Crystal structure of the Triticum aestivum xylanase inhibitor-I in complex with aspergillus niger xylanase-I | | Descriptor: | Endo-1,4-beta-xylanase I, GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
1FIZ
 
 | | THREE DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM BOAR SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
1M9J
 
 | | human endothelial nitric oxide synthase with chlorzoxazone bound | | Descriptor: | CHLORZOXAZONE, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1FQA
 
 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3C
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (E132C, E133G, R191G mutant) from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1FB7
 
 | | CRYSTAL STRUCTURE OF AN IN VIVO HIV-1 PROTEASE MUTANT IN COMPLEX WITH SAQUINAVIR: INSIGHTS INTO THE MECHANISMS OF DRUG RESISTANCE | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, HIV-1 PROTEASE | | Authors: | Hong, L, Zhang, X.C, Hartsuck, J.A, Tang, J. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance.
Protein Sci., 9, 2000
|
|
1FBU
 
 | | HEAT SHOCK TRANSCRIPTION FACTOR DNA BINDING DOMAIN | | Descriptor: | HEAT SHOCK FACTOR PROTEIN | | Authors: | Hardy, J.A, Nelson, H.C.M. | | Deposit date: | 2000-07-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor.
Protein Sci., 9, 2000
|
|
3P2R
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate | | Descriptor: | Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
1M8I
 
 | | inducible nitric oxide synthase with 5-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
3PDI
 
 | | Precursor bound NifEN | | Descriptor: | IRON/SULFUR CLUSTER, L-Cluster (Fe8S9), Nitrogenase MoFe cofactor biosynthesis protein NifE, ... | | Authors: | Kaiser, J.T, Hu, Y, Wiig, J.A, Rees, D.C, Ribbe, M.W. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Science, 331, 2011
|
|
1M8E
 
 | | inducible nitric oxide synthase with 7-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
