8RN0
 
 | |
8RN4
 
 | |
8RN5
 
 | |
8RN9
 
 | |
8RMP
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core1) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN1
 
 | | Influenza B polymerase, monomeric encapsidase with 5' cRNA hook bound | | Descriptor: | 5' cRNA hook (1-12), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RNA
 
 | | Influenza B polymerase apo-trimer | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RMR
 
 | |
8RN3
 
 | |
8RMQ
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core2) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RNB
 
 | |
8RMS
 
 | |
8RN7
 
 | |
8PSN
 
 | |
8PTJ
 
 | |
8QZ8
 
 | |
1EAJ
 
 | |
1F5W
 
 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
1H7Z
 
 | | Adenovirus Ad3 fibre head | | Descriptor: | ADENOVIRUS FIBRE PROTEIN, SULFATE ION | | Authors: | Durmort, C, Stehlin, C, Schoehn, G, Mitraki, A, Drouet, E, Cusack, S, Burmeister, W.P. | | Deposit date: | 2001-01-21 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Fiber Head of Ad3, a Non-Car-Binding Serotype of Adenovirus
Virology, 285, 2001
|
|
1H6K
 
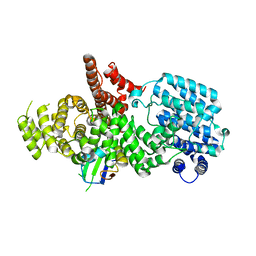 | | nuclear Cap Binding Complex | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, CBP80 | | Authors: | Mazza, C, Ohno, M, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Nuclear CAP Binding Complex
Mol.Cell, 8, 2001
|
|
4CQN
 
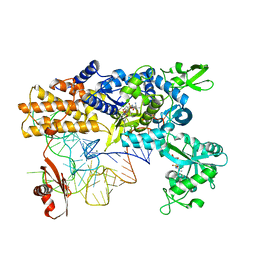 | | Crystal structure of the E.coli LeuRS-tRNA complex with the non- cognate isoleucyl adenylate analogue | | Descriptor: | ESCHERICHIA COLI TRNA-LEU UAA ISOACCEPTOR, LEUCINE--TRNA LIGASE, MAGNESIUM ION, ... | | Authors: | Palencia, A, Cusack, S, Cvetesic, N, Haslaz, I, Gruic-Sovulj, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Physiological Target for Leurs Translational Quality Control is Norvaline
Embo J., 33, 2014
|
|
3ZGZ
 
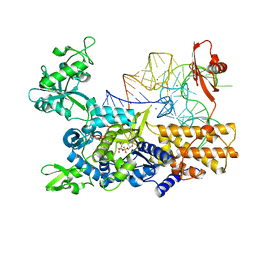 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
4CGS
 
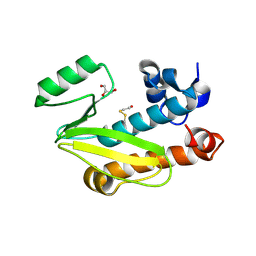 | | Crystal structure of the N-terminal domain of the PA subunit of Dhori virus polymerase | | Descriptor: | GLYCEROL, POLYMERASE SUBUNIT PA | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-26 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CHF
 
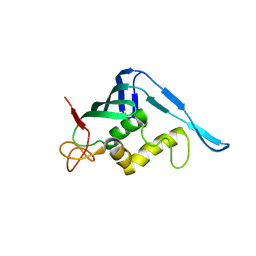 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase (form 2) | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CHD
 
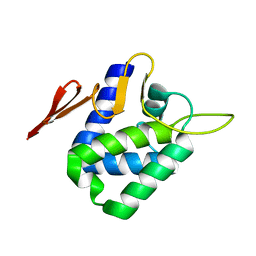 | | Crystal structure of the '627' domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
