3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
4HQQ
 
 | | Crystal structure of RV144-elicited antibody CH58 | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain | | Authors: | McLellan, J.S, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
3NKE
 
 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
5VDU
 
 | |
5VDV
 
 | |
1JZT
 
 | |
4L4W
 
 | |
5IDI
 
 | | Structure of beta glucosidase 1A from Thermotoga neapolitana, mutant E349A | | Descriptor: | 1,4-beta-D-glucan glucohydrolase, ACETATE ION | | Authors: | Kulkarni, T, Nordberg Karlsson, E, Logan, D.T. | | Deposit date: | 2016-02-24 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-glucosidase 1A from Thermotoga neapolitana and comparison of active site mutants for hydrolysis of flavonoid glucosides.
Proteins, 85, 2017
|
|
2IVS
 
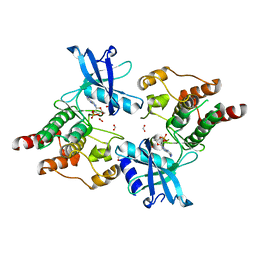 | | Crystal structure of non-phosphorylated RET tyrosine kinase domain | | Descriptor: | 2',3'- cyclic AMP, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Chemical Inhibition of the Ret Tyrosine Kinase Domain.
J.Biol.Chem., 281, 2006
|
|
2H2Q
 
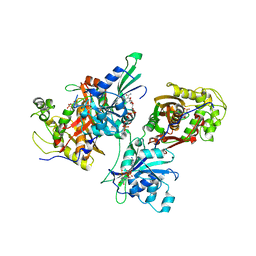 | | Crystal structure of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Senkovich, O, Schormann, N, Chattopadhyay, D. | | Deposit date: | 2006-05-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3G9L
 
 | |
3G9N
 
 | |
3NS7
 
 | |
4HPY
 
 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | Descriptor: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
5T7V
 
 | | Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Belousoff, M.J, Lithgow, T, Eyal, Z, Yonath, A, Radjainia, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-05-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in theStaphylococcus aureusRibosome.
MBio, 8, 2017
|
|
2IVT
 
 | |
4HPO
 
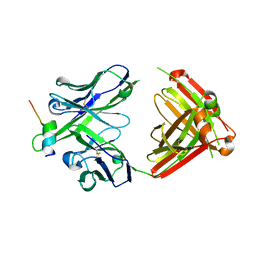 | | Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
2IVV
 
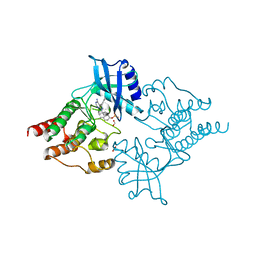 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
2IVU
 
 | |
7DNF
 
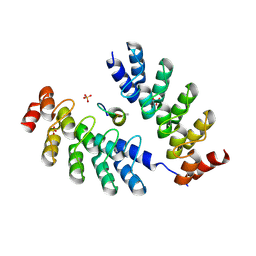 | | DARPin 63_B7 in complex with V3-IY (MN) crown mimetic | | Descriptor: | DARPin 63_B7, SULFATE ION, V3-IY (MN) crown mimetic peptide | | Authors: | Wu, Y, Plueckthun, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7DNG
 
 | |
7DNE
 
 | |
1BI2
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
6IUV
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 3C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3C11 Heavy Chain, 3C11 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
7EKM
 
 | | Mitochondrial outer membrane protein | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Zhang, S.S. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular insights into the human ABCB6 transporter.
Cell Discov, 7, 2021
|
|
