7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7XME
 
 | | Structure of Influenza A virus polymerase basic protein 2 (PB2) with an azazindole derivative | | Descriptor: | (2~{S},3~{S})-3-[[5-dimethoxyphosphoryl-4-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, IODIDE ION, Polymerase basic protein 2 | | Authors: | Zhang, Z. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Discovery of a novel azaindole derivatives targeting the influenza PB2 cap binding region
To Be Published
|
|
7YTP
 
 | | TLR7 in complex with an inhibitor | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2022-08-15 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | TLR7 in complex with an inhibitor
To Be Published
|
|
3VAX
 
 | | Crystal structure of DndA from streptomyces lividans | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein dndA | | Authors: | Zhang, Z, Chen, F, Lin, K, Wu, G. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation
Plos One, 7, 2012
|
|
1TPB
 
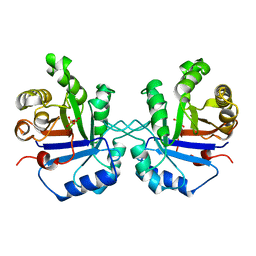 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1TPC
 
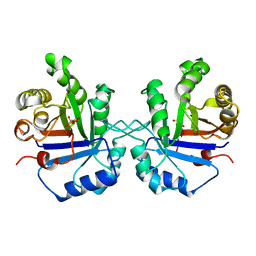 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
6YPC
 
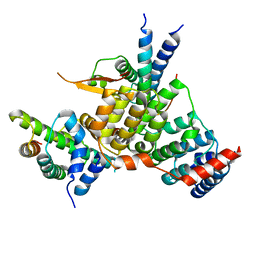 | | Crystal structure of the kinetochore subunits H/I/K/T/W penta-complex from S. cerevisiae at 2.9 angstroms | | Descriptor: | Inner kinetochore subunit CNN1, Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, ... | | Authors: | Bellini, D, Zhang, Z, Barford, D. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Cenp-HIKHead-TW sub-module of the inner kinetochore CCAN complex.
Nucleic Acids Res., 48, 2020
|
|
1TPU
 
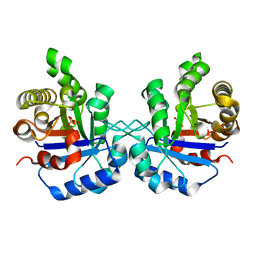 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
5JO4
 
 | | Antibody Fab Fragment Complex | | Descriptor: | D80 Fab Heavy Chain, D80 Fab Light Chain, G6 Fab Heavy Chain, ... | | Authors: | Zhang, Z, Prachanronarong, K.P, Marasco, W.A, Schiffer, C.A.S. | | Deposit date: | 2016-05-01 | | Release date: | 2017-11-08 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
5JQD
 
 | | Antibody Fab Fragment | | Descriptor: | D80 Fab Fragment Heavy Chain, D80 Fab Fragment Light Chain | | Authors: | Zhang, Z, Prachanronarong, K, Gellatly, K, Marasco, W.A, Schiffer, C.A. | | Deposit date: | 2016-05-04 | | Release date: | 2017-11-08 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
4XPU
 
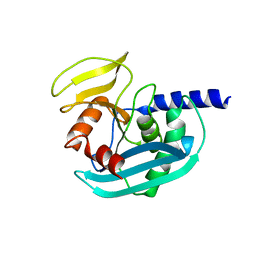 | | The crystal structure of EndoV from E.coli | | Descriptor: | Endonuclease V | | Authors: | Xie, W, Zhang, Z. | | Deposit date: | 2015-01-18 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of E. coli endonuclease V, an essential enzyme for deamination repair
Sci Rep, 5, 2015
|
|
5WRO
 
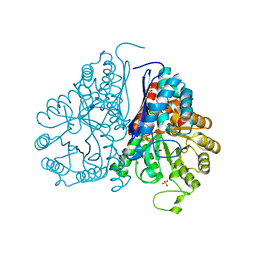 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8ZMS
 
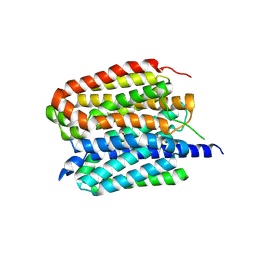 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
8ZMR
 
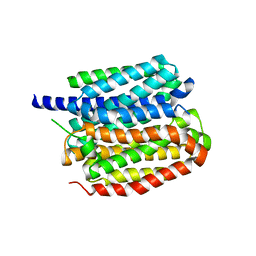 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
4NSP
 
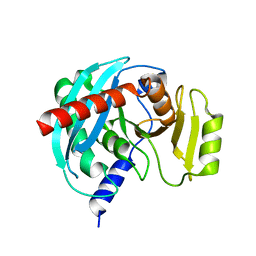 | | Crystal structure of human ENDOV | | Descriptor: | Endonuclease V | | Authors: | Xie, W, Zhang, Z, Hao, Z. | | Deposit date: | 2013-11-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human endonuclease V as an inosine-specific ribonuclease.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3KAE
 
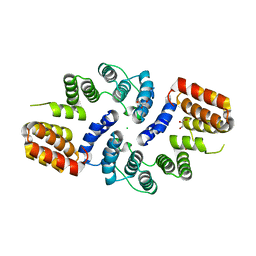 | | Cdc27 N-terminus | | Descriptor: | CHLORIDE ION, GLYCEROL, Possible protein of nuclear scaffold, ... | | Authors: | Barford, D, Zhang, Z, Roe, S.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture
J.Mol.Biol., 397, 2010
|
|
3PX7
 
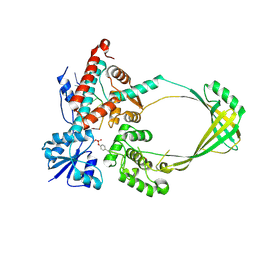 | |
8WH6
 
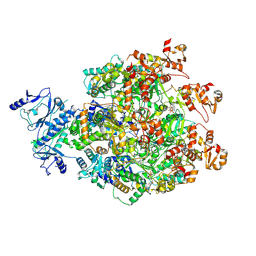 | |
8WH4
 
 | | MPOX E5 hexamer ssDNA bound apo conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WH3
 
 | | MPOX E5 hexamer apo form | | Descriptor: | Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
5MOF
 
 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form in complex with manganese and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, CHLORIDE ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LUN
 
 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - P1 ultra-high resolution crystal form in complex with iron, N-oxalylglycine and arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, FE (III) ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8Y3X
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Dong, H, Chen, Y. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
