1PR6
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-xylofuranosyladenine and Phosphate/Sulfate | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR5
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 7-deazaadenosine and Phosphate/Sulfate | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR0
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Inosine and Phosphate/Sulfate | | Descriptor: | INOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR2
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-[2-deoxyribofuranosyl]-6-methylpurine and Phosphate/Sulfate | | Descriptor: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR1
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Formycin B and Phosphate/Sulfate | | Descriptor: | FORMYCIN B, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1U2Y
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
1EER
 
 | |
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
8D1T
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5DWZ
 
 | | Structural and functional characterization of PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-(Acyl carrier protein) synthase III, 3-oxoacyl-[acyl-carrier-protein] synthase 3, ... | | Authors: | Drees, S.L, Li, C, Prasetya, F, Saleem, M, Dreveny, I, Hennecke, U, Williams, P, Emsley, J, Fetzner, S. | | Deposit date: | 2015-09-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM.
J.Biol.Chem., 291, 2016
|
|
4PQW
 
 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
1Z23
 
 | | The serine-rich domain from Crk-associated substrate (p130Cas) | | Descriptor: | CRK-associated substrate | | Authors: | Briknarova, K, Nasertorabi, F, Havert, M.L, Eggleston, E, Hoyt, D.W, Li, C, Olson, A.J, Vuori, K, Ely, K.R. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle.
J.Biol.Chem., 280, 2005
|
|
7C7Q
 
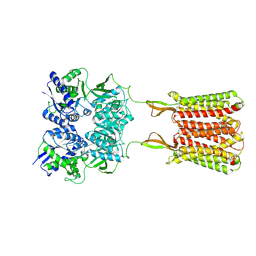 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
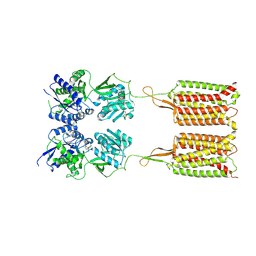 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
4Q3H
 
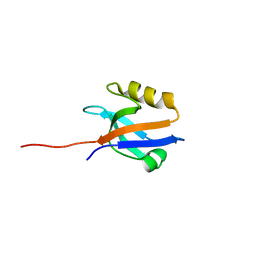 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
1YSW
 
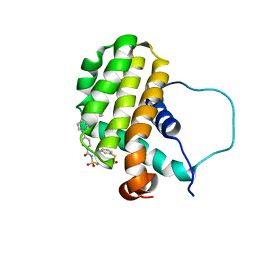 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSG
 
 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1ZYW
 
 | | Crystal Structure Of Mutant K8DP9SR58KP60G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZUT
 
 | | Crystal Structure Of Mutant K8DP9SR58K Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
6UGH
 
 | |
1ZYV
 
 | | Crystal Structure Of Mutant K8DP9SR58KV59G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-12 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
