1YVG
 
 | | Structural analysis of the catalytic domain of tetanus neurotoxin | | Descriptor: | Tetanus toxin, light chain, ZINC ION | | Authors: | Rao, K.N, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the catalytic domain of tetanus neurotoxin.
Toxicon, 45, 2005
|
|
1ZKW
 
 | | Crystal structure of Arg347Ala mutant of botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
3UHJ
 
 | | Crystal structure of a probable glycerol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Probable glycerol dehydrogenase, SELENIUM ATOM, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a probable glycerol dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
2MFP
 
 | |
3WBD
 
 | | Crystal structure of anti-polysialic acid antibody single chain Fv fragment (mAb735) complexed with octasialic acid | | Descriptor: | CITRATE ANION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, single chain Fv fragment of mAb735 | | Authors: | Nagae, M, Ikeda, A, Hanashima, S, Kitajima, K, Sato, C, Yamaguchi, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: insight into the binding preference for polysialic acid.
J.Biol.Chem., 288, 2013
|
|
3TXV
 
 | | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021 | | Descriptor: | Probable tagatose 6-phosphate kinase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021
To be Published
|
|
2OG9
 
 | |
2OGK
 
 | |
4M1A
 
 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
2NWU
 
 | |
2OPJ
 
 | |
2OOD
 
 | |
2OQH
 
 | |
2P0L
 
 | |
2OY9
 
 | |
2P9B
 
 | |
2PBZ
 
 | |
3T4Z
 
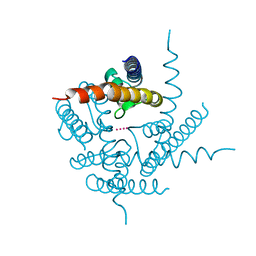 | | Crystal Structure of NaK2K Channel Y55W Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4IMZ
 
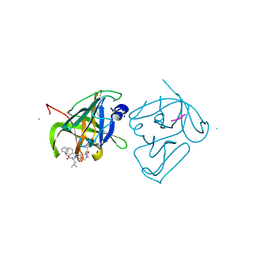 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
3U5R
 
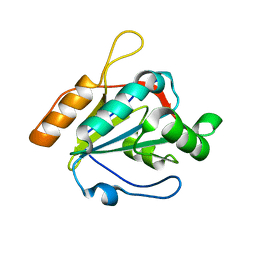 | |
6JBF
 
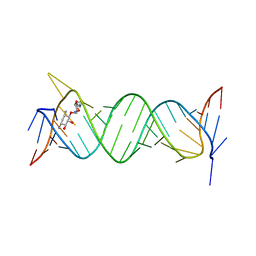 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
6JBG
 
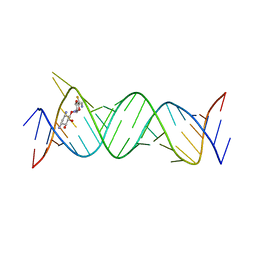 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (equatorial 4'-F) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-glucopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
3SQS
 
 | |
3TCU
 
 | | Crystal Structure of NaK2K Channel D68E Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
