6OXO
 
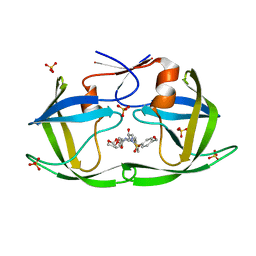 | | HIV-1 Protease NL4-3 WT in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
8FLC
 
 | |
6OXP
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXS
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-76 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
8FLD
 
 | |
5NBW
 
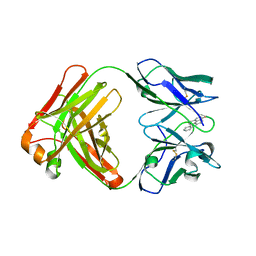 | |
8CAT
 
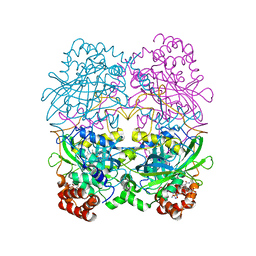 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
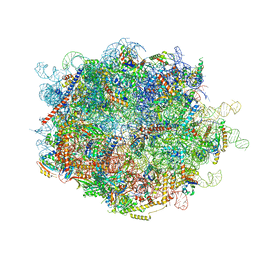
5MRC
 
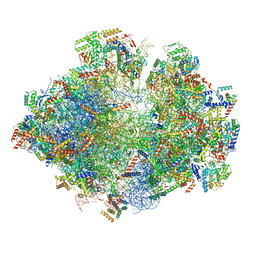 | | Structure of the yeast mitochondrial ribosome - Class A | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
2KZ8
 
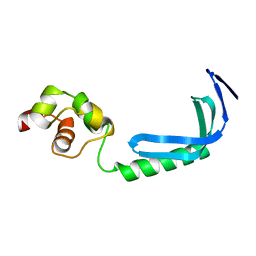 | | Solution NMR structure of MqsA, a protein from E. coli, containing a Zinc finger, N-terminal and a Helix Turn-Helix C-terminal domain | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygiT | | Authors: | Papadopoulos, E, Vlamis-Gardikas, A, Graslund, A, Billeter, M, Holmgren, A, Collet, J. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biophysical properties of MqsA, a Zn-containing antitoxin from Escherichia coli
Biochim.Biophys.Acta, 2012
|
|
3RSM
 
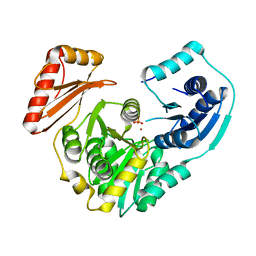 | | Crystal structure of S108C mutant of PMM/PGM | | Descriptor: | PHOSPHATE ION, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Akella, A, Anbanandam, A, Kelm, A, Wei, Y, Mehra-Chaudhary, R, Beamer, L, Van Doren, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect.
Biochemistry, 51, 2012
|
|
6QNQ
 
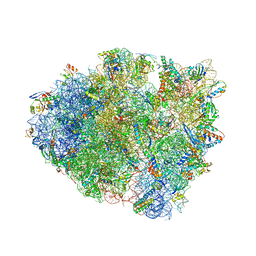 | | 70S ribosome initiation complex (IC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
1O83
 
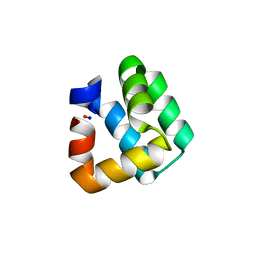 | | Crystal Structure of Bacteriocin AS-48 at pH 7.5, phosphate bound. Crystal form I | | Descriptor: | GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, PHOSPHATE ION | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Valdivia, E, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-25 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
1H4S
 
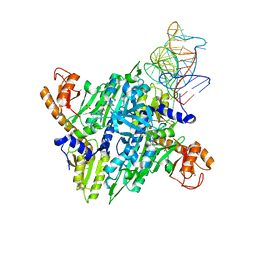 | | Prolyl-tRNA synthetase from Thermus thermophilus complexed with tRNApro(CGG) and a prolyl-adenylate analogue | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, PROLYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Succession of Substrate Induced Conformational Changes Ensures the Amino Acid Specificity of Thermus Thermophilus Prolyl-tRNA Synthetase: Comparison with Histidyl-tRNA Synthetase
J.Mol.Biol., 309, 2001
|
|
6ZTL
 
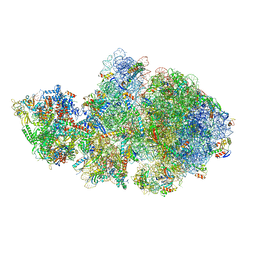 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
2O9A
 
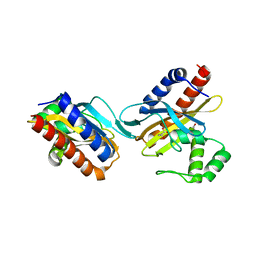 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
1GUZ
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1GYX
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
5D7U
 
 | | Crystal structure of the C-terminal domain of MMTV integrase | | Descriptor: | ISOPROPYL ALCOHOL, Pr160 | | Authors: | Cook, N.J, Pye, V.E, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-08-14 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
6ZS8
 
 | |
6O50
 
 | | Binary complex of native hAChE with BW284c51 | | Descriptor: | 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
8H2N
 
 | |
6QDT
 
 | | Crystal structure of 14-3-3sigma in complex with a RapGef2 pT740 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
