2BJI
 
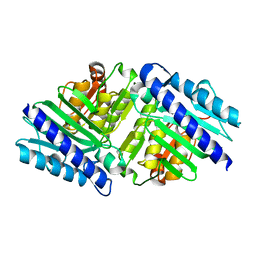 | | High Resolution Structure of myo-Inositol Monophosphatase, The Target of Lithium Therapy | | Descriptor: | INOSITOL-1(OR 4)-MONOPHOSPHATASE, MAGNESIUM ION | | Authors: | Gill, R, Mohammed, F, Badyal, R, Coates, L, Erskine, P, Thompson, D, Cooper, J, Gore, M, Wood, S. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
2BJN
 
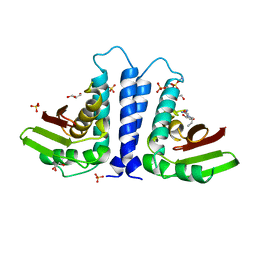 | | X-ray Structure of human TPC6 | | Descriptor: | GLYCEROL, SULFATE ION, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 6B | | Authors: | Kummel, D, Mueller, J.J, Roske, Y, Misselwitz, R, Bussow, K, Heinemann, U. | | Deposit date: | 2005-02-04 | | Release date: | 2005-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Trapp Subunit Tpc6 Suggests a Model for a Trapp Subcomplex.
Embo Rep., 6, 2005
|
|
2BKW
 
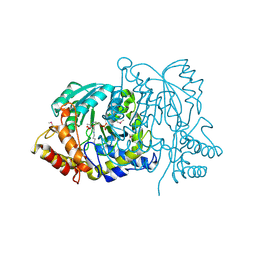 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
2BNK
 
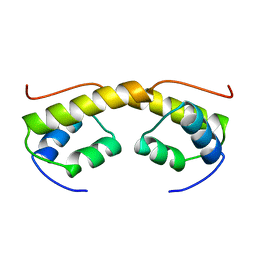 | | The structure of phage phi29 replication organizer protein p16.7 | | Descriptor: | EARLY PROTEIN GP16.7 | | Authors: | Albert, A, Asensio, J.L, Munoz-Espin, D, Gonzalez, C, Hermoso, J.A, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-03-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Functional Domain of {Varphi}29 Replication Organizer: Insights Into Oligomerization and DNA Binding.
J.Biol.Chem., 280, 2005
|
|
2BUD
 
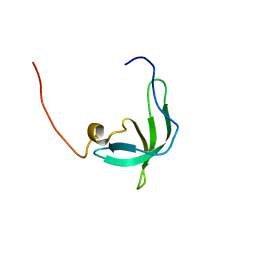 | | The solution structure of the chromo barrel domain from the males- absent on the first (MOF) protein | | Descriptor: | MALES-ABSENT ON THE FIRST PROTEIN | | Authors: | Nielsen, P.R, Nietlispach, D, Buscaino, A, Warner, R.J, Akhtar, A, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Chromo Barrel Domain from the Mof Acetyltransferase
J.Biol.Chem., 280, 2005
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
2C5B
 
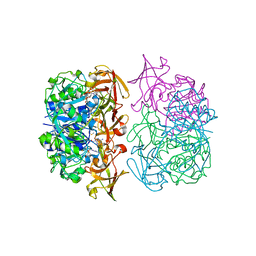 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
2BOK
 
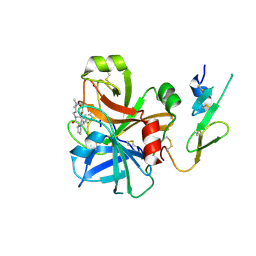 | | Factor Xa - cation | | Descriptor: | COAGULATION FACTOR X, SODIUM ION, [AMINO (4-{(3AS,4R,8AS,8BR)-1,3-DIOXO-2- [3-(TRIMETHYLAMMONIO) PROPYL]DECAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL}PHENYL) METHYLENE]AMMONIUM | | Authors: | Morgenthaler, M, Schaerer, K, Paulini, R, Obst-Sander, U, Banner, D.W, Schlatter, D, Benz, J, Stihle, M, Diederich, F. | | Deposit date: | 2005-04-12 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Quantification of Cation-Pi Interactions in Protein-Ligand Complexes: Crystal-Structure Analysis of Factor Xa Bound to a Quaternary Ammonium Ion Ligand
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2BYO
 
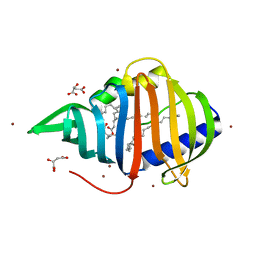 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
2BDX
 
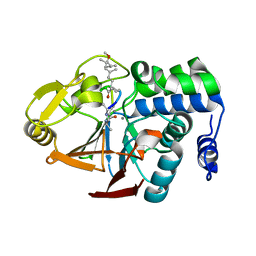 | | X-ray Crystal Structure of dihydromicrocystin-LA bound to Protein Phosphatase-1 | | Descriptor: | DIHYDROMICROCYSTIN-LA, MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-gamma catalytic subunit | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-21 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2BZM
 
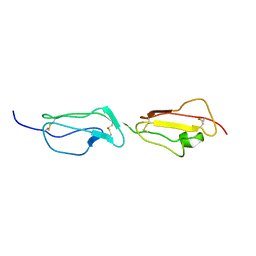 | | Solution structure of the primary host recognition region of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Uhrin, D, Lyon, M, Pangburn, M.K, Barlow, P.N. | | Deposit date: | 2005-08-18 | | Release date: | 2006-03-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Disease-Associated Sequence Variations Congregate in a Polyanion Recognition Patch on Human Factor H Revealed in Three-Dimensional Structure.
J.Biol.Chem., 281, 2006
|
|
2BKQ
 
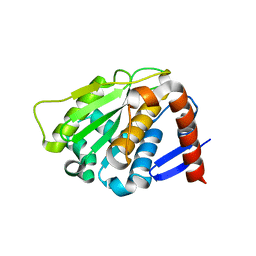 | | NEDD8 protease | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 8 | | Authors: | Shen, L.N, Liu, H, Dong, C, Xirodimas, D, Naismith, J.H, Hay, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Nedd8 Ubiquitin Discrimination by the Deneddylating Enzyme Nedp1
Embo J., 24, 2005
|
|
2C4N
 
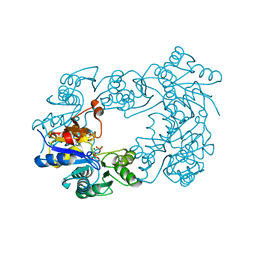 | | NagD from E.coli K-12 strain | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PROTEIN NAGD | | Authors: | Tremblay, L.W, Dunaway-Mariano, D, Allen, K. | | Deposit date: | 2005-10-20 | | Release date: | 2006-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Activity Analyses of Escherichia Coli K-12 Nagd Provide Insight Into the Evolution of Biochemical Function in the Haloalkanoic Acid Dehalogenase Superfamily
Biochemistry, 45, 2006
|
|
2C4U
 
 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2C5X
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, Mcinnes, C, Pandalaneni, S.R, Mcnae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2BF0
 
 | | crystal structure of the rpr of pcf11 | | Descriptor: | CALCIUM ION, PCF11 | | Authors: | Noble, C.G, Hollingworth, D, Martin, S.R, Adeniran, V.E, Smerdon, S.J, Kelly, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2004-12-02 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key Features of the Interaction between Pcf11 Cid and RNA Polymerase II Ctd.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BN8
 
 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | Descriptor: | CELL DIVISION ACTIVATOR CEDA | | Authors: | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | Deposit date: | 2005-03-22 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
5SMN
 
 | | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework -- Crystal structure of SARS-CoV-2 main protease in complex with Z1365651030 (Mpro-IBM0078) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(1-cyanocyclopropyl)-1-(3-methylpyridin-4-yl)piperidine-4-carboxamide | | Authors: | Fearon, D, Owen, C.D, Lukacik, P, Strain-Damerell, C.M, von Delft, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework
To Be Published
|
|
4PPR
 
 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
8CBD
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
5SML
 
 | | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework -- Crystal structure of SARS-CoV-2 main protease in complex with Z68337194 (Mpro-IBM0045) | | Descriptor: | 3C-like proteinase, 6-{[(3,4-dichlorophenyl)methyl](methyl)amino}pyridine-3-sulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Lukacik, P, Strain-Damerell, C.M, von Delft, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework
To Be Published
|
|
5SMM
 
 | | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework -- Crystal structure of SARS-CoV-2 main protease in complex with Z1633315555 (Mpro-IBM0058) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[4-(3-fluorophenyl)oxan-4-yl]-2-(3-hydroxyphenyl)acetamide | | Authors: | Fearon, D, Owen, C.D, Lukacik, P, Strain-Damerell, C.M, von Delft, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework
To Be Published
|
|
4PST
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 277 K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PTJ
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 277K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
8CCK
 
 | |
