5FOE
 
 | | Crystal structure of the C. elegans Protein O-fucosyltransferase 2 (CePOFUT2) double mutant (R298K-R299K) in complex with GDP and the human TSR1 from thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 2,Thrombospondin-1, ... | | Authors: | Valero-Gonzalez, J, Leonhard-Melief, C, Lira-Navarrete, E, Jimenez-Oses, G, Hernandez-Ruiz, C, Pallares, M.C, Yruela, I, Vasudevan, D, Lostao, A, Corzana, F, Takeuchi, H, Haltiwanger, R.S, Hurtado-Guerrero, R. | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Proactive Role of Water Molecules in Acceptor Recognition of Thrombospondin Type 1 Repeats by Protein-O-Fucosyltransferase 2
Nat.Chem.Biol., 12, 2016
|
|
5FQ2
 
 | |
8UOU
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
8UTA
 
 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | Descriptor: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
5FW8
 
 | | Structure of human transthyretin mutant E89K | | Descriptor: | TRANSTHYRETIN | | Authors: | Gallego, P, Varejao, N, Santanna, R, Saraiva, M.J, Ventura, S, Reverter, D. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of New Non-Amyloidogenic Mutant of Ttr A108V
To be Published
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
5FYA
 
 | | Cubic crystal of the native PlpD | | Descriptor: | PATATIN-LIKE PROTEIN, PLPD | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
1CDR
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
5G0I
 
 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker cleaved) in complex with Nexturastat A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HDAC6, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
8UXN
 
 | | Caulobacter crescentus FljM flagellar filament (symmetrized) | | Descriptor: | Flagellin FljM | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
8VD6
 
 | |
8UXJ
 
 | | Caulobacter crescentus FljK flagellar filament (asymmetrical) | | Descriptor: | Flagellin FljK | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Atomic-level architecture of Caulobacter crescentus flagellar filaments provide evidence for multi-flagellin filament stabilization
To Be Published
|
|
8VAC
 
 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAE
 
 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
5GAE
 
 | | RNC in complex with a translocating SecYEG | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-25 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
5GVS
 
 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
8VAF
 
 | | Cryogenic electron microscopy structure of apo human serum albumin | | Descriptor: | Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
4JGK
 
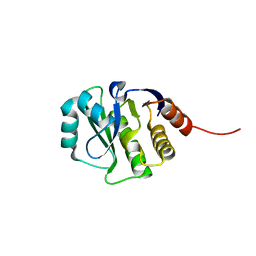 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
5GIB
 
 | | Succinic acid bound trypsin crystallized as dimer | | Descriptor: | CALCIUM ION, Cationic trypsin, SUCCINIC ACID | | Authors: | Kutumbarao, N.H.V, Manohar, R, KarthiK, L, Malathy, P, Gunasekaran, K, Velmurugan, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Trypsin bound with Succinic acid
To Be Published
|
|
5GKM
 
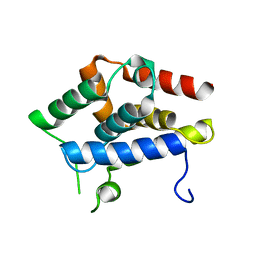 | |
6GYP
 
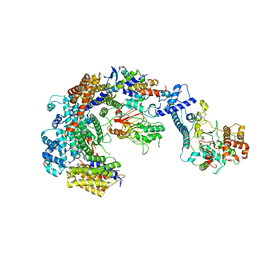 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5GVR
 
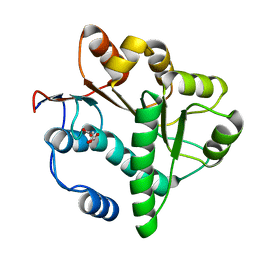 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
7CLU
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
