6YTF
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
371D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
6Z1M
 
 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
3NVE
 
 | |
3A31
 
 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix (selenomethionine derivative) | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
2E4Z
 
 | | Crystal structure of the ligand-binding region of the group III metabotropic glutamate receptor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metabotropic glutamate receptor 7 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1LPU
 
 | | Low Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
3A3Z
 
 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277A(C23S) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-2-methyl-9,10-secoandrosta-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
2JM1
 
 | |
3AAU
 
 | | Bovine beta-trypsin bound to meta-diguanidino schiff base copper (II) chelate | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cationic trypsin, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
3OWN
 
 | | Potent macrocyclic renin inhibitors | | Descriptor: | (2S,4S)-4-hydroxy-2-(1-methylethyl)-4-[(4R,13S)-18-[methyl(methylsulfonyl)amino]-2,15-dioxo-4-phenyl-11-oxa-3,14-diazatricyclo[14.3.1.1~5,9~]henicosa-1(20),5(21),6,8,16,18-hexaen-13-yl]-N-(2-methylpropyl)butanamide, (2S,4S)-4-hydroxy-2-(1-methylethyl)-4-[(4S,13S)-18-[methyl(methylsulfonyl)amino]-2,15-dioxo-4-phenyl-11-oxa-3,14-diazatricyclo[14.3.1.1~5,9~]henicosa-1(20),5(21),6,8,16,18-hexaen-13-yl]-N-(2-methylpropyl)butanamide, ACETATE ION, ... | | Authors: | Borkakoti, N, Derbyshire, D. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent macrocyclic renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3A78
 
 | | Crystal structure of the human VDR ligand binding domain bound to the natural metabolite 1alpha,25-dihydroxy-3-epi-vitamin D3 | | Descriptor: | (1S,3S,5Z,7E,14beta,17alpha)-9,10-secocholesta-5,7,10-triene-1,3,25-triol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Sigueiro, R, Antony, P, Rochel, N, Moras, D. | | Deposit date: | 2009-09-18 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the activation of the human Vitamin D receptor by the natural metabolite 1alpha,25-dihydroxy-3-epi-vitamin D3
To be Published
|
|
370D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
382D
 
 | |
1YVE
 
 | | ACETOHYDROXY ACID ISOMEROREDUCTASE COMPLEXED WITH NADPH, MAGNESIUM AND INHIBITOR IPOHA (N-HYDROXY-N-ISOPROPYLOXAMATE) | | Descriptor: | ACETOHYDROXY ACID ISOMEROREDUCTASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Biou, V, Dumas, R, Cohen-Addad, C, Douce, R, Job, D, Pebay-Peyroula, E. | | Deposit date: | 1996-10-11 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of plant acetohydroxy acid isomeroreductase complexed with NADPH, two magnesium ions and a herbicidal transition state analog determined at 1.65 A resolution.
EMBO J., 16, 1997
|
|
4IRB
 
 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
1TVT
 
 | |
369D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1TOF
 
 | | THIOREDOXIN H (OXIDIZED FORM), NMR, 23 STRUCTURES | | Descriptor: | THIOREDOXIN H | | Authors: | Mittard, V, Blackledge, M.J, Stein, M, Jacquot, J.-P, Marion, D, Lancelin, J.-M. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii.
Eur.J.Biochem., 243, 1997
|
|
383D
 
 | |
3OZR
 
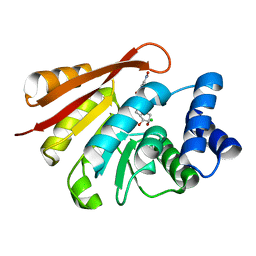 | | Rat catechol O-methyltransferase in complex with a catechol-type, bisubstrate inhibitor, no substituent in the adenine site - humanized form | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
2E2J
 
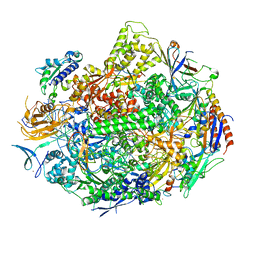 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
7P84
 
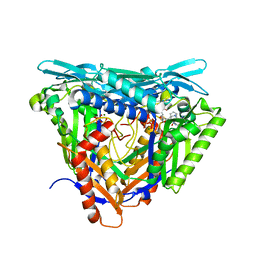 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P8M
 
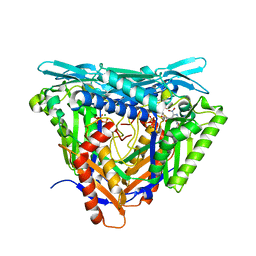 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with DMNB-SAM (4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | 4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine, MAGNESIUM ION, S-adenosylmethionine synthase, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
2EB4
 
 | | Crystal structure of apo-HpcG | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, SODIUM ION, THIOCYANATE ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
