3FXU
 
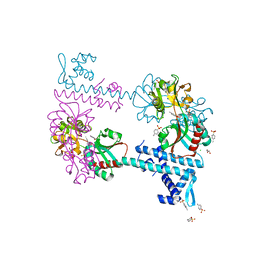 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
1AAM
 
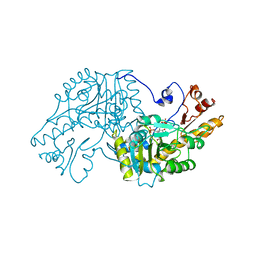 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
6OJ0
 
 | | Cryo-EM reconstruction of Sulfolobus polyhedral virus 1 (SPV1) | | Descriptor: | Structural protein VP4, Uncharacterized protein | | Authors: | Wang, F, Liu, Y, Conway, J.F, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A packing for A-form DNA in an icosahedral virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7D68
 
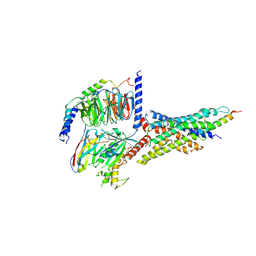 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
2HGA
 
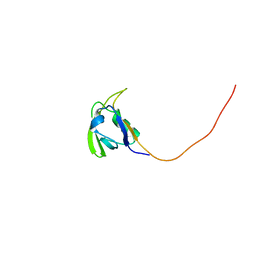 | | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A | | Descriptor: | Conserved protein MTH1368 | | Authors: | Liu, G, Lin, Y, Parish, D, Shen, Y, Sukumaran, D, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A
TO BE PUBLISHED
|
|
6OBJ
 
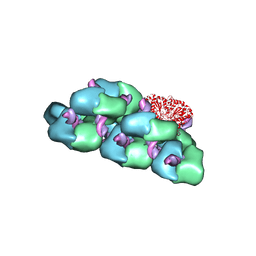 | |
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6OEX
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor 3-(2-{1-[2-(Piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3- thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine | | Descriptor: | 3-(2-{1-[2-(piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OPH
 
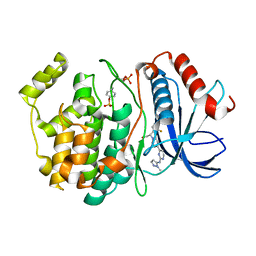 | | phosphorylated ERK2 with GDC-0994 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1C7F
 
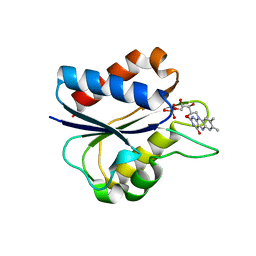 | | D95E OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
2IRP
 
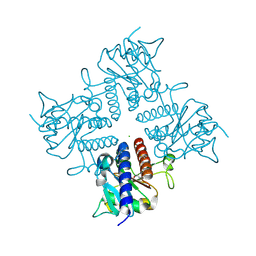 | | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Putative aldolase class 2 protein aq_1979 | | Authors: | Jeyakanthan, J, Gayathri, D, Yogavel, M, Velmurugan, D, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5
To be Published
|
|
3FR1
 
 | |
6HIW
 
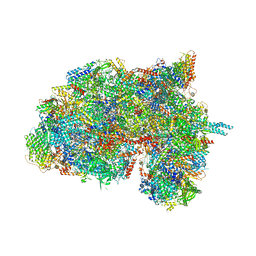 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete small mitoribosomal subunit in complex with mt-IF-3 | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ramrath, D, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-09-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
6OTC
 
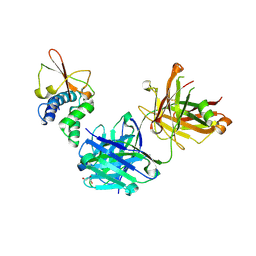 | | Synthetic Fab bound to Marburg virus VP35 interferon inhibitory domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Polymerase cofactor VP35, ... | | Authors: | Amatya, P, Chen, G, Borek, D, Sidhu, S.S, Leung, D.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Marburg Virus RNA Synthesis by a Synthetic Anti-VP35 Antibody.
Acs Infect Dis., 5, 2019
|
|
6OAO
 
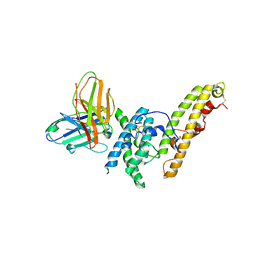 | | Structure of DBP in complex with human neutralizing antibody 092096 | | Descriptor: | Antibody 092096 single chain variable fragment, Duffy binding surface protein region II, SULFATE ION | | Authors: | Urusova, D, Tolia, N.H. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.497 Å) | | Cite: | Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP.
Nat Microbiol, 4, 2019
|
|
3OWS
 
 | |
6O6A
 
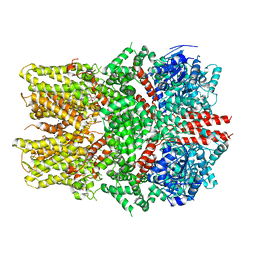 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, ligand-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
1ADU
 
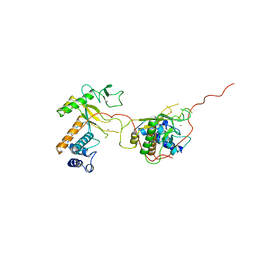 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
6O7U
 
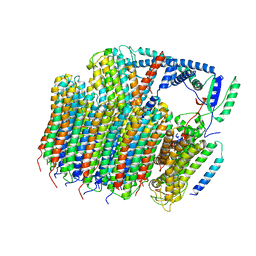 | | Saccharomyces cerevisiae V-ATPase Stv1-VO | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit a, Golgi isoform, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1A3B
 
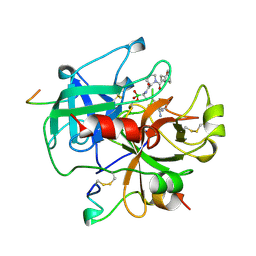 | | COMPLEX OF HUMAN ALPHA-THROMBIN WITH THE BIFUNCTIONAL BORONATE INHIBITOR BOROLOG1 | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), Hirudin, ... | | Authors: | Skordalakes, E, Elgendy, S, Dodson, G, Goodwin, C.A, Green, D, Scully, M.F, Freyssinet, J.H, Kakkar, V.V, Deadman, J. | | Deposit date: | 1998-01-20 | | Release date: | 1998-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional peptide boronate inhibitors of thrombin: crystallographic analysis of inhibition enhanced by linkage to an exosite 1 binding peptide.
Biochemistry, 37, 1998
|
|
3FTK
 
 | |
3FVA
 
 | | NNQNTF segment from elk prion | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2009-01-15 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3PA9
 
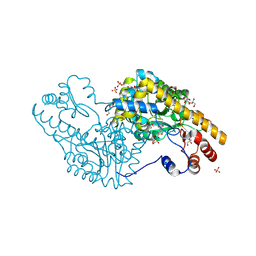 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
6OFD
 
 | | The crystal structure of octadecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P, Kan, M.W. | | Deposit date: | 2019-03-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
