2XB7
 
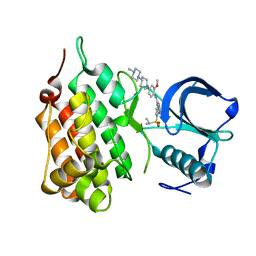 | | Structure of Human Anaplastic Lymphoma Kinase in complex with NVP- TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, ALK TYROSINE KINASE RECEPTOR | | Authors: | Bossi, R.T, Saccardo, M.B, Ardini, E, Menichincheri, M, Rusconi, L, Magnaghi, P, Orsini, P, Fogliatto, G, Bertrand, J.A. | | Deposit date: | 2010-04-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Anaplastic Lymphoma Kinase in Complex with ATP Competitive Inhibitors.
Biochemistry, 49, 2010
|
|
2VSL
 
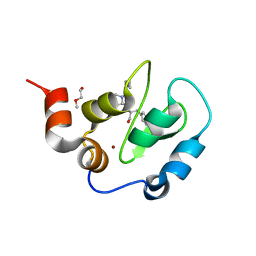 | | Crystal Structure of XIAP BIR3 with a Bivalent Smac Mimetic | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, PEPTIDE (MAA-LYS-PRO-PHE), POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interaction of a Cyclic, Bivalent Smac Mimetic with the X-Linked Inhibitor of Apoptosis Protein.
Biochemistry, 47, 2008
|
|
4ZNK
 
 | | Thermus Phage P74-26 Large Terminase ATPase domain from (P 32 2 1 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3H8L
 
 | | The first X-ray structure of a sulfide:quinone oxidoreductase: insights into sulfide oxidation mechanism | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase, PHOSPHATE ION, ... | | Authors: | Brito, J.A, Sousa, F.L, Stelter, M, Bandeiras, T.M, Vonrhein, C, Teixeira, M, Pereira, M.M, Archer, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into sulfide:quinone oxidoreductase.
Biochemistry, 48, 2009
|
|
3HBP
 
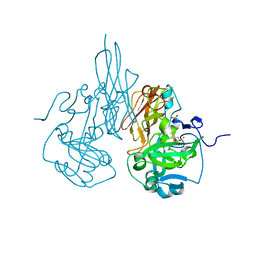 | | The crystal structure of C185S mutant of recombinant sulfite oxidase with bound substrate, sulfite, at the active site | | Descriptor: | HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFITE ION, ... | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
2W35
 
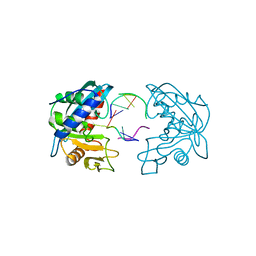 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W2D
 
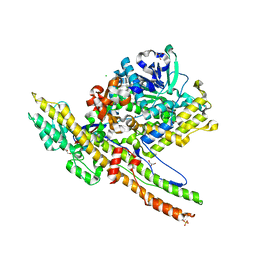 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3GLX
 
 | | Crystal Structure Analysis of the DtxR(E175K) complexed with Ni(II) | | Descriptor: | Diphtheria toxin repressor, NICKEL (II) ION, PHOSPHATE ION | | Authors: | D'Aquino, J.A, Denninger, A, Moulin, A, D'Aquino, K.E, Ringe, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Decreased sensitivity to changes in the concentration of metal ions as the basis for the hyperactivity of DtxR(E175K).
J.Mol.Biol., 390, 2009
|
|
2VWT
 
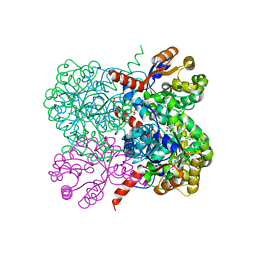 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
5A52
 
 | |
3GYH
 
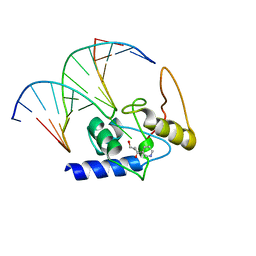 | | Crystal Structure Analysis of S. Pombe ATL in complex with damaged DNA containing POB | | Descriptor: | 1-PYRIDIN-3-YLBUTAN-1-ONE, Alkyltransferase-like protein 1, DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A, Shin, D.S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
2W22
 
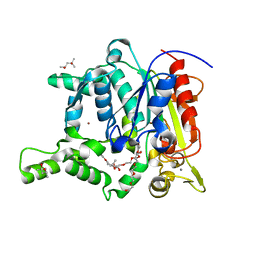 | | Activation Mechanism of Bacterial Thermoalkalophilic Lipases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Carrasco-Lopez, C, Godoy, C, De Las Rivas, B, Fernandez-Lorente, G, Palomo, J.M, Guisan, J.M, Fernandez-Lafuente, R, Hermoso, J.A. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements.
J. Biol. Chem., 284, 2009
|
|
5AG4
 
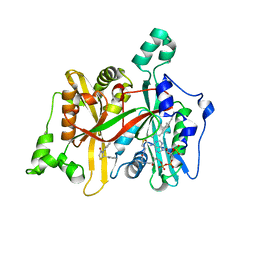 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2S)-3-(3-chlorophenyl)-2-(pyridin-2-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
2VR9
 
 | |
3GZF
 
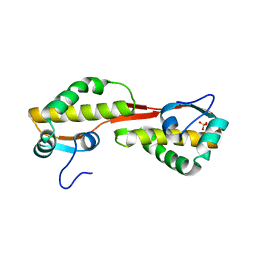 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4ZV6
 
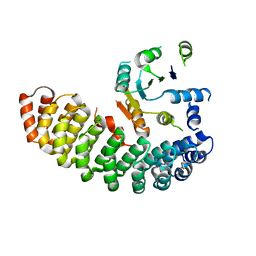 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
3GIS
 
 | |
2VWS
 
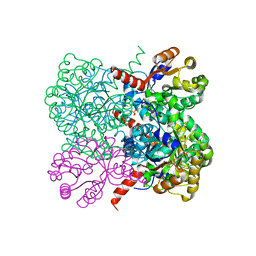 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 | | Descriptor: | GLYCEROL, PHOSPHATE ION, YFAU, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
3GJC
 
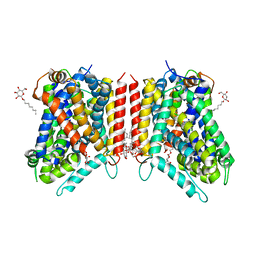 | | Crystal Structure of the E290S mutant of LeuT with bound OG | | Descriptor: | LEUCINE, SODIUM ION, Transporter, ... | | Authors: | Winther, A.M.L, Quick, M, Javitch, J.A, Nissen, P. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W43
 
 | | Structure of L-haloacid dehalogenase from S. tokodaii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL 2-HALOALKANOIC ACID DEHALOGENASE, PHOSPHATE ION | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
2VYQ
 
 | | FERREDOXIN:NADP REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY, ALA 160 REPLACED BY THR, LEU 263 REPLACED BY PRO AND TYR 303 REPLACED BY SER (T155G-A160T-L263P-Y303S) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Peregrina, J.R, Medina, M. | | Deposit date: | 2008-07-28 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein Motifs Involved in Coenzyme Interaction and Enzymatic Efficiency in Anabaena Ferredoxin-Nadp+ Reductase.
Biochemistry, 48, 2009
|
|
3GP6
 
 | | Crystal structure of PagP in SDS/MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DODECYL SULFATE, ... | | Authors: | Cuesta-Seijo, J.A, Prive, G.G. | | Deposit date: | 2009-03-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler.
Structure, 18, 2010
|
|
3GVA
 
 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
2WAJ
 
 | | Crystal structure of human Jnk3 complexed with a 1-aryl-3,4- dihydroisoquinoline inhibitor | | Descriptor: | 1-(3-BROMOPHENYL)-7-CHLORO-6-METHOXY-3,4-DIHYDROISOQUINOLINE, MITOGEN-ACTIVATED PROTEIN KINASE 10 | | Authors: | Bax, B.D, Christopher, J.A, Jones, E.J, Mosley, J.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1-Aryl-3,4-Dihydroisoquinoline Inhibitors of Jnk3.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
