8XG2
 
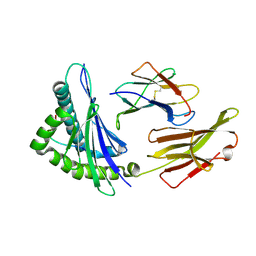 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
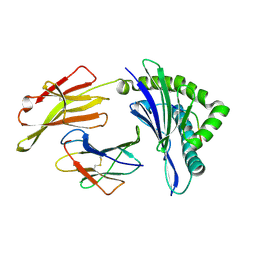 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
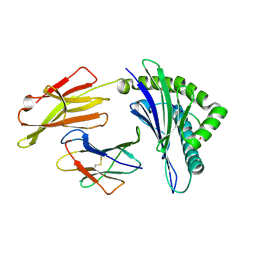 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
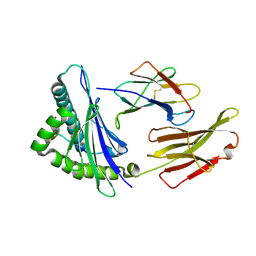 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
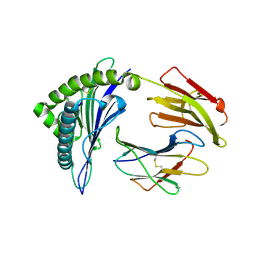 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
5YZV
 
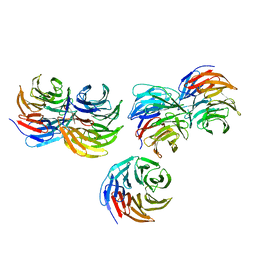 | | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA | | Descriptor: | Probable serine/threonine-protein kinase PkwA | | Authors: | Li, D.Y, Shen, C, Du, Y, Qiao, F.F, Kong, T, Yuan, L.R, Zhang, D.L, Wu, X.H, Wu, Y.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA
Sci Rep, 8, 2018
|
|
7KHL
 
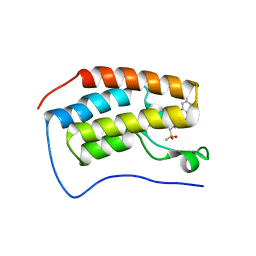 | | BRD4-BD1 Compound6 (methyl 4-(3,5-difluoropyridin-2-yl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxylate) | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, methyl 7-(3,5-difluoropyridin-2-yl)-2-methyl-10-[(methylsulfonyl)methyl]-3-oxo-3,4,6,7-tetrahydro-2H-2,4,7-triazadibenzo[cd,f]azulene-9-carboxylate | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.286 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
7KHH
 
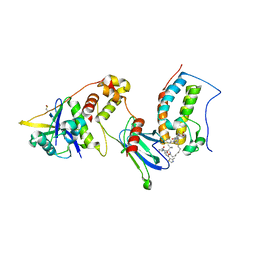 | | Ternary complex of VHL/BRD4-BD1/Compound9 (4-(3,5-difluoropyridin-2-yl)-N-(11-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-11-oxoundecyl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxamide) | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
8J5D
 
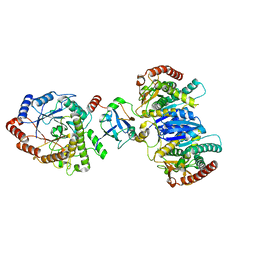 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
8JGT
 
 | |
8JGQ
 
 | |
8JGO
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGR
 
 | |
8JGX
 
 | |
8JGP
 
 | |
6M79
 
 | |
6MC3
 
 | |
6MC2
 
 | |
5XKX
 
 | | Crystal structure of WT DddY | | Descriptor: | ACETATE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5Y4K
 
 | | Crystal structure of DddY mutant Y260A | | Descriptor: | ACRYLIC ACID, DMSP lyase DddY, ZINC ION | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2017-08-03 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
6N4G
 
 | |
5XN5
 
 | |
6J2E
 
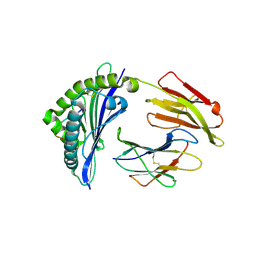 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP1 | | Descriptor: | Beta-2-microglobulin, EBOV-NP1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
