7NG3
 
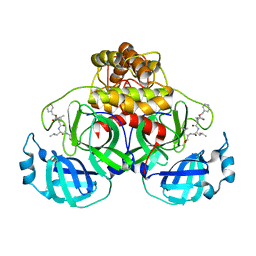 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NG6
 
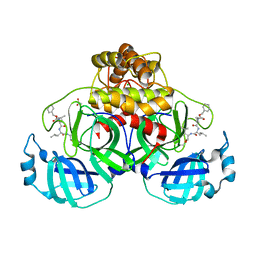 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NF5
 
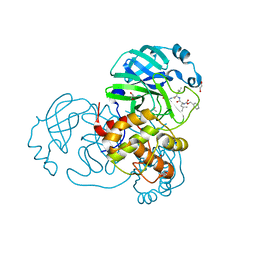 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8ZCL
 
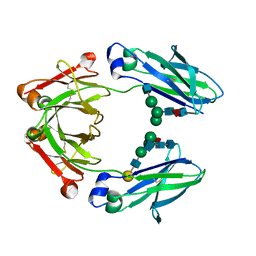 | | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
8ZCK
 
 | | Serial Femtosecond Crystallography Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial Femtosecond Crystallography Structure of Fc Fragment of Human IgG1 from VEGF-Trap
To Be Published
|
|
8ZCM
 
 | | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
9IIE
 
 | | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
3ZY1
 
 | |
1VE0
 
 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
7BE7
 
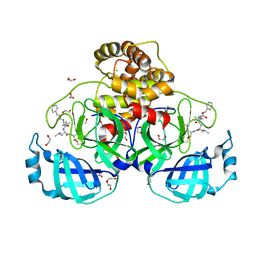 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BGP
 
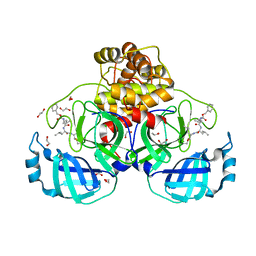 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BB2
 
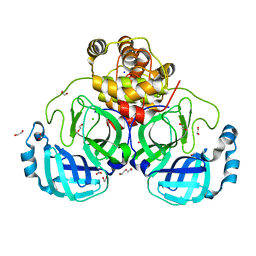 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALI
 
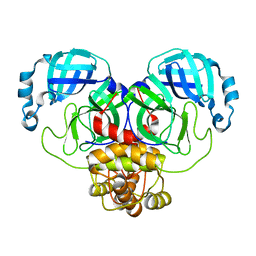 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NBY
 
 | | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 5-nitro-1,3-thiazole, CHLORIDE ION, Main Protease, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
7BFB
 
 | | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, Main Protease, N-phenyl-2-selanylbenzamide, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
1EID
 
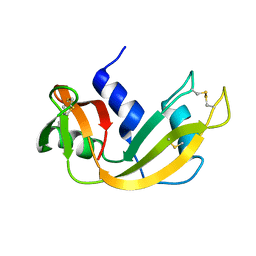 | |
1EIC
 
 | |
1EIE
 
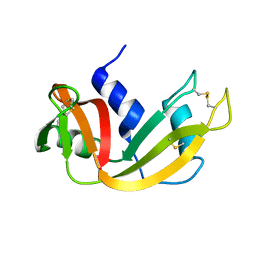 | |
2O43
 
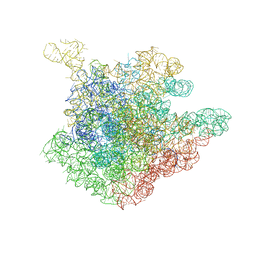 | | Structure of 23S rRNA of the large ribosomal subunit from Deinococcus radiodurans in complex with the macrolide erythromycylamine | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-10-AMINO-6-{[(2S,3R,4S,6R)-4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PY RAN-2-YL]OXY}-14-ETHYL-7,12,13-TRIHYDROXY-4-{[(2R,4R,5S,6S)-5-HYDROXY-4-METHOXY-4,6-DIMETHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY }-3,5,7,9,11,13-HEXAMETHYLOXACYCLOTETRADECAN-2-ONE, 23S rRNA | | Authors: | Pyetan, E, Baram, D, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2006-12-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Chemical parameters influencing fine tuning in the binding of Macrolide antibiotics to the ribosomal tunnel
TO BE PUBLISHED
|
|
1FS3
 
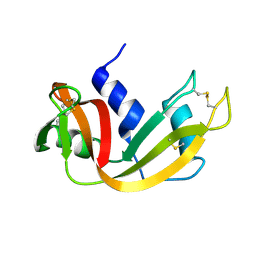 | |
2O44
 
 | | Structure of 23S rRNA of the large ribosomal subunit from Deinococcus radiodurans in complex with the macrolide josamycin | | Descriptor: | (2S,3S,4R,6S)-6-{[(2R,3S,4R,5R,6S)-6-{[(4R,5S,6S,7R,9R,10S,12E,14Z,16R)-4-(ACETYLOXY)-10-HYDROXY-5-METHOXY-9,16-DIMETHYL-2-OXO-7-(2-OXOETHYL)OXACYCLOHEXADECA-12,14-DIEN-6-YL]OXY}-4-(DIMETHYLAMINO)-5-HYDROXY-2-METHYLTETRAHYDRO-2H-PYRAN-3-YL]OXY}-4-HYDROXY-2,4-DIMETHYLTETRAHYDRO-2H-PYRAN-3-YL 3-METHYLBUTANOATE, 23S rRNA | | Authors: | Pyetan, E, Daram, D, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2006-12-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chemical parameters influencing fine tuning in the binding of Macrolide antibiotics to the ribosomal tunnel
TO BE PUBLISHED
|
|
2M1C
 
 | | HADDOCK structure of GtYybT PAS Homodimer | | Descriptor: | DHH subfamily 1 protein | | Authors: | Liang, Z.X, Pervushin, K, Tan, E, Rao, F, Pasunooti, S, Soehano, I, Lescar, J. | | Deposit date: | 2012-11-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PAS Domain of a Thermophilic YybT Protein Homolog Reveals a Potential Ligand-binding Site.
J.Biol.Chem., 288, 2013
|
|
8VIW
 
 | | Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site | | Descriptor: | Heparosan synthase B, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Krahn, J.M, Pedersen, L.C, Liu, J, Stancanelli, E, Borgnia, M, Vivarette, E. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
