2A15
 
 | | X-ray Crystal Structure of RV0760 from Mycobacterium Tuberculosis at 1.68 Angstrom Resolution | | Descriptor: | HYPOTHETICAL PROTEIN Rv0760c, NICOTINAMIDE | | Authors: | Garen, C.R, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase
Biochim.Biophys.Acta, 1784, 2008
|
|
2I3A
 
 | | Crystal structure of N-Acetyl-gamma-Glutamyl-Phosphate Reductase (Rv1652) from Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Moraidin, F, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of N-acetyl-gamma-glutamyl-phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+).
J.Mol.Biol., 367, 2007
|
|
2I3G
 
 | | Crystal structure of N-Acetyl-gamma-Glutamyl-Phosphate Reductase (Rv1652) from Mycobacterium tuberculosis in complex with NADP+. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-gamma-glutamyl-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Moraidin, F, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of N-acetyl-gamma-glutamyl-phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+).
J.Mol.Biol., 367, 2007
|
|
2GJX
 
 | | Crystallographic structure of human beta-Hexosaminidase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase alpha chain, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic structure of human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss
of GM2 Ganglioside Hydrolysis
J.Mol.Biol., 359, 2006
|
|
2A4O
 
 | | Dual modes of modification of Hepatitis A virus 3C protease by a serine derived beta-lactone: selective crytstallization and high resolution structure of the His102 adduct | | Descriptor: | ACETYL GROUP, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PHENYLALANINE AMIDE, ... | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2JEK
 
 | | Crystal structure of the conserved hypothetical protein Rv1873 from Mycobacterium tuberculosis at 1.38 A | | Descriptor: | GLYCEROL, RV1873, SULFATE ION | | Authors: | Garen, C.R, Cherney, M.M, Bergmann, E.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Molecular Structure of Rv1873, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis, at 1.38A Resolution
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2D41
 
 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2CXV
 
 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
2DJ5
 
 | | Crystal Structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Cherney, M.M, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2NU0
 
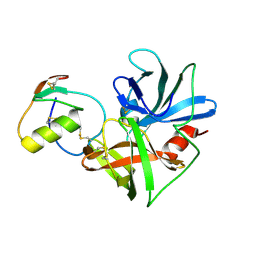 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I
To be Published
|
|
2O2H
 
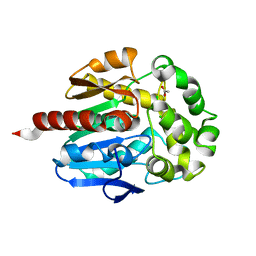 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane | | Descriptor: | 1,2-DICHLOROETHANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane
TO BE PUBLISHED
|
|
2NU3
 
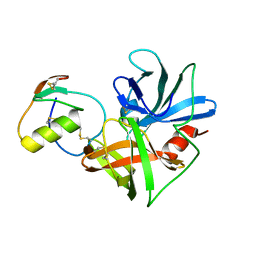 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2O2I
 
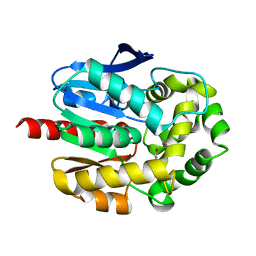 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol
To be Published
|
|
2NU2
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2NU1
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
To be Published
|
|
2NR9
 
 | | Crystal structure of GlpG, Rhomboid Peptidase from Haemophilus influenzae | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, Protein glpG homolog | | Authors: | Lemieux, M.J, Fischer, S.J, Cherney, M.M, Bateman, K.S, James, M.N.G. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NU4
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1MBQ
 
 | | Anionic Trypsin from Pacific Chum Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, Trypsin | | Authors: | Toyota, E, Ng, K.K.S, Kuninaga, S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Nucleotide Sequence of an Anionic Trypsin from Chum Salmon (Oncorhynchus keta) in Comparison with Atlantic Salmon (Salmo salar) and Bovine Trypsin
J.Mol.Biol., 324, 2002
|
|
1MBM
 
 | | NSP4 proteinase from Equine Arteritis Virus | | Descriptor: | chymotrypsin-like serine protease | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Mark, B.L, van Aken, D, Cherney, M.M, Garen, C, Kolodenko, Y, Gorbalenya, A.E, Snijder, E.J, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Arterivirus nsp4: the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole
J.Biol.Chem., 277, 2002
|
|
1NHV
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
