4R76
 
 | |
4R5T
 
 | |
4R6T
 
 | |
7N4S
 
 | | Bruton's tyrosine kinase in complex with compound 65 | | Descriptor: | (3R,3'R)-3-anilino-1'-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)[1,3'-bipiperidin]-2-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
7N4R
 
 | | Bruton's tyrosine kinase in complex with compound 21 | | Descriptor: | DIMETHYL SULFOXIDE, N-{2-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]ethyl}-N~2~-phenylglycinamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
7N4Q
 
 | | Bruton's tyrosine kinase in complex with compound 45 | | Descriptor: | (2R)-2-(3-chloro-5-fluoroanilino)-2-cyclopropyl-N-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]acetamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
8UAK
 
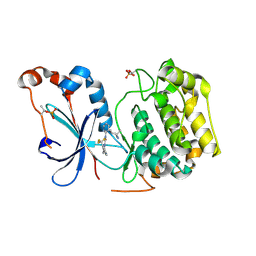 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
3L9M
 
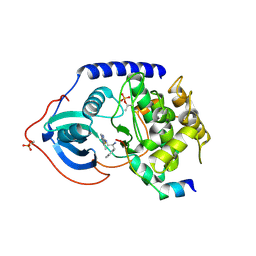 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9N
 
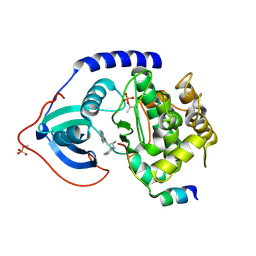 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7SPT
 
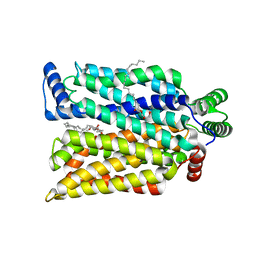 | | Crystal structure of exofacial state human glucose transporter GLUT3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
7SPS
 
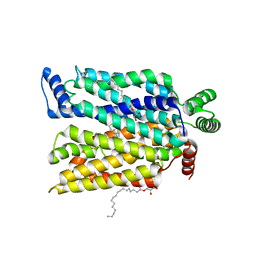 | | Crystal structure of human glucose transporter GLUT3 bound with exofacial inhibitor SA47 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
3L9L
 
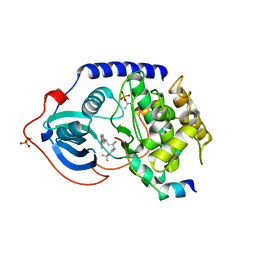 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8SAI
 
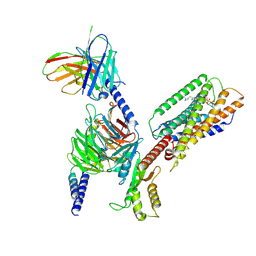 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7T17
 
 | |
7WHB
 
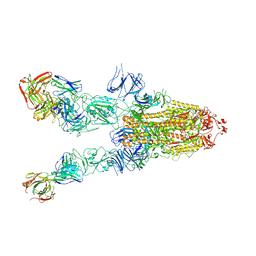 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
7WH8
 
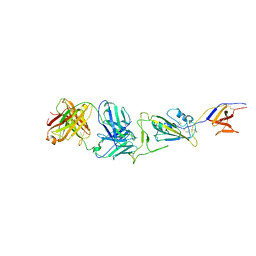 | |
7WHD
 
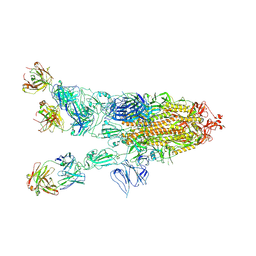 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
4U1D
 
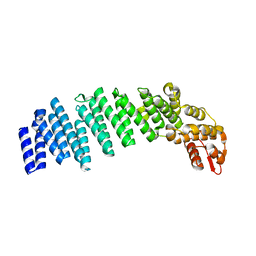 | |
2G25
 
 | | E. Coli Pyruvate Dehydrogenase Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
4U1C
 
 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
2G28
 
 | | E. Coli Pyruvate Dehydrogenase H407A variant Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
7V21
 
 | |
7V1Y
 
 | |
7V1Z
 
 | | human Serine beta-lactamase-like protein LACTB | | Descriptor: | Serine beta-lactamase-like protein LACTB, mitochondrial | | Authors: | Zhang, M.H, Yang, M.J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB.
Structure, 30, 2022
|
|
