6C3E
 
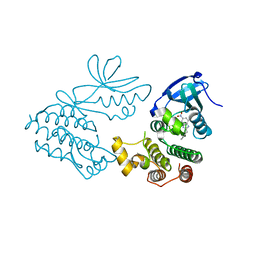 | | CRYSTAL STRUCTURE OF RIP1 KINASE BOUND TO INHIBITOR | | Descriptor: | 2-benzyl-5-nitro-1H-benzimidazole, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
6C4D
 
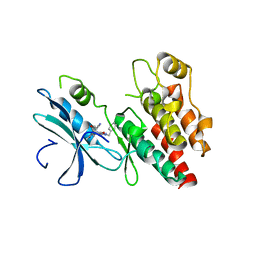 | | Structure based design of RIP1 kinase inhibitors | | Descriptor: | (3S)-3-(2-benzyl-3-chloro-7-oxo-2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepine-8-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
6MAU
 
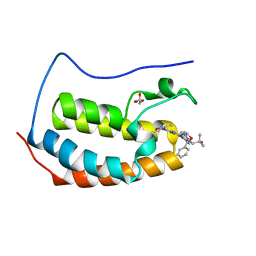 | | Crystal structure of human BRD4(1) in complex with CN210 (compound 19) | | Descriptor: | 1-(4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-[(3S)-3-phenylmorpholin-4-yl]quinazolin-2-yl}-1H-pyrazol-1-yl)-2-methylpropan-2-ol, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Nadupalli, A, Fontano, E, Connors, C.R, Chan, S.G, Olland, A.M, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
1MUP
 
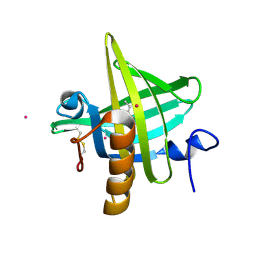 | | PHEROMONE BINDING TO TWO RODENT URINARY PROTEINS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN | | Authors: | Bocskei, Z, Flower, D.R, Groom, C.R, Phillips, S.E.V, North, A.C.T. | | Deposit date: | 1992-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography.
Nature, 360, 1992
|
|
7A9K
 
 | |
7A9F
 
 | |
7A9M
 
 | |
3D03
 
 | | 1.9A structure of Glycerophoshphodiesterase (GpdQ) from Enterobacter aerogenes | | Descriptor: | COBALT (II) ION, Phosphohydrolase | | Authors: | Hadler, K.S, Tanifum, E, Yip, S.H.-C, Miti, N, Guddat, L.W, Jackson, C.J, Gahan, L.R, Carr, P.D, Nguyen, K, Ollis, D.L, Hengge, A.C, Larrabee, J.A, Schenk, G. | | Deposit date: | 2008-04-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-promoted formation of a catalytically competent binuclear center and regulation of reactivity in a glycerophosphodiesterase from Enterobacter aerogenes.
J.Am.Chem.Soc., 130, 2008
|
|
2YBG
 
 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4BY5
 
 | |
4BY4
 
 | |
2D4F
 
 | | The Crystal Structure of human beta2-microglobulin | | Descriptor: | Beta-2-microglobulin, SODIUM ION | | Authors: | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
2D4D
 
 | | The Crystal Structure of human beta2-microglobulin, L39W W60F W95F Mutant | | Descriptor: | Beta-2-microglobulin, SODIUM ION | | Authors: | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
7DUH
 
 | |
7DUG
 
 | |
7DUK
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''MS bound | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 30S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H, Destan, E. | | Deposit date: | 2021-01-09 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''MS bound
to be published
|
|
7DUI
 
 | |
7DUJ
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''Bz bound | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 30S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H, Destan, E. | | Deposit date: | 2021-01-09 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''Bz bound
to be published
|
|
2WQJ
 
 | |
2WQI
 
 | |
2WTT
 
 | |
2OPO
 
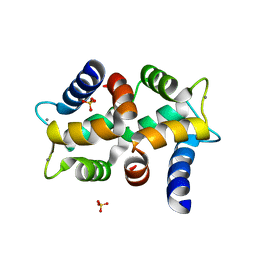 | | Crystal structure of the calcium-binding pollen allergen Che a 3 | | Descriptor: | CALCIUM ION, Polcalcin Che a 3, SULFATE ION | | Authors: | Verdino, P, Keller, W. | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structure of the cross-reactive pollen allergen Che a 3: visualizing cross-reactivity on the molecular surfaces of weed, grass, and tree pollen allergens.
J.Immunol., 180, 2008
|
|
