4AQ5
 
 | |
6KFY
 
 | |
6KG1
 
 | | NifS from Helicobacter pylori, soaked with L-cysteine for 180 sec | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
3WV4
 
 | | Crystal structure of VinN | | Descriptor: | Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
 | | Complex structure of VinN with L-aspartate | | Descriptor: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-30 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WD7
 
 | | Type III polyketide synthase | | Descriptor: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WD8
 
 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
6M0Q
 
 | | Hydroxylamine oxidoreductase from Nitrosomonas europaea | | Descriptor: | Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
6M0P
 
 | | Hydroxylamine oxidoreductase in complex with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
3WI3
 
 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | Descriptor: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2013-09-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
4CDO
 
 | | Crystal structure of PQBP1 bound to spliceosomal U5-15kD | | Descriptor: | THIOREDOXIN-LIKE PROTEIN 4A, POLYGLUTAMINE-BINDING PROTEIN | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | Authors: | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1VA3
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
5XHJ
 
 | | Crystal Structure of P450BM3 with 5-Cyclohexylvaleroyl-L-Tryptophan | | Descriptor: | 5-cyclohexylpentanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suzuki, K, Shoji, O, Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of stereoselectivity of benzylic hydroxylation catalysed by wild-type cytochrome P450BM3 using decoy molecules
CATALYSIS SCIENCE AND TECHNOLOGY, 7, 2017
|
|
5YK9
 
 | |
5YHJ
 
 | | Cytochrome P450EX alpha (CYP152N1) wild-type with myristic acid | | Descriptor: | Cytochrome P450, MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Onoda, H, Shoji, O, Suzuki, K, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-09-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-Oxidative Decarboxylation of Fatty Acids Catalysed by Cytochrome P450 Peroxygenases Yielding Shorter-Alkyl-Chain Fatty Acids
Catalysis Science And Technology, 2017
|
|
7E15
 
 | |
1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
2LX2
 
 | | 1H,13C,15N assignments for an isoform of the type III antifreeze protein from notched-fin eelpout | | Descriptor: | Type III antifreeze protein nfeAFP11 | | Authors: | Kumeta, H, Ogura, K, Nishimiya, Y, Miura, A, Inagaki, F, Tsuda, S. | | Deposit date: | 2012-08-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: a defective isoform and its activity-improved variant of a type III antifreeze protein from Zoarces elongates Kner
J.Biomol.Nmr, 55, 2013
|
|
5WT5
 
 | | L-homocysteine-bound NifS from Helicobacter pylori | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshot of cysteine desulfurase NifS with L-cysteine in initiation of catalysis
to be published
|
|
5X9A
 
 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
5XV8
 
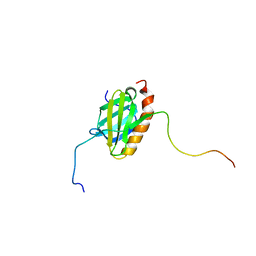 | |
5XT6
 
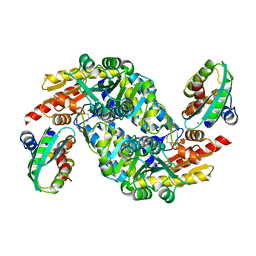 | | A sulfur-transferring catalytic intermediate of SufS-SufU complex from Bacillus subtilis | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Cysteine desulfurase SufS, ZINC ION, ... | | Authors: | Fujishiro, T, Kunichika, K, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
5XT5
 
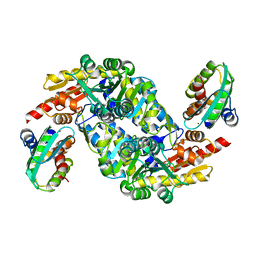 | | SufS-SufU complex from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
