1JTP
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, LYSOZYME C, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JUJ
 
 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
7QUF
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CK156 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-~{tert}-butyl-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8DG7
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
5U29
 
 | | Crystal structure of Cryptococcus neoformans H99 Acetyl-CoA Synthetase in complex with Ac-AMS | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(acetylsulfamoyl)adenosine, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Edwards, T.E, Potts, K.T, Taylor, B.M. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cryptococcus neoformans H99 Acetyl-CoA Synthetase in complex with Ac-AMS
To Be Published
|
|
8DGJ
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGI
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8DG5
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
7QF2
 
 | | Structure of miniSOG reconstituted with riboflavin as a cofactor | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
5TYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
4MC9
 
 | | HIV protease in complex with AA74 | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
7QF4
 
 | | Structure of the R57Q mutant of miniSOG expressed in E. coli in LB medium enriched with riboflavin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, RIBOFLAVIN, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
7QF3
 
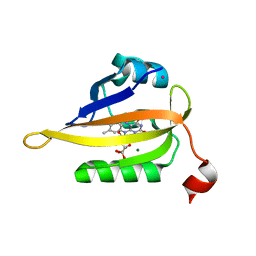 | | Structure of the R57Q mutant of miniSOG expressed in E. coli in regular LB medium | | Descriptor: | CHLORIDE ION, COBALT (II) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
5U8T
 
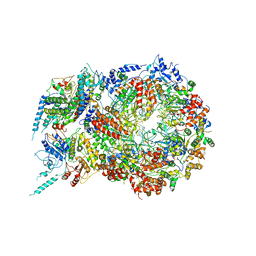 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UBD
 
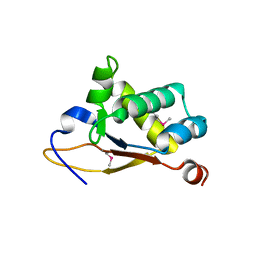 | | Crystal structure of the N-terminal domain (domain 1) of RctB, RctB-1-124-L48M | | Descriptor: | RctB replication initiator protein | | Authors: | Orlova, N, Ivashkiv, O, Waldor, M.K, Jeruzalmi, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The replication initiator of the cholera pathogen's second chromosome shows structural similarity to plasmid initiators.
Nucleic Acids Res., 45, 2017
|
|
7QF5
 
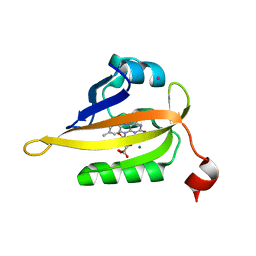 | | Structure of the Q103L mutant of miniSOG | | Descriptor: | CHLORIDE ION, COBALT (II) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4PR9
 
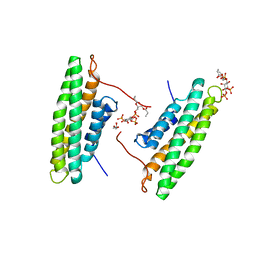 | | Human Vinculin (residues 891-1066) in complex with PIP | | Descriptor: | GLYCEROL, Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Rangarajan, E.S, Patil, D, Izard, T. | | Deposit date: | 2014-03-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism and function of lipid-directed oligomerization of vinculin
To be Published
|
|
7QHB
 
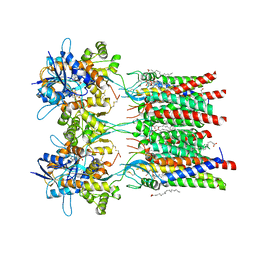 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
8DBA
 
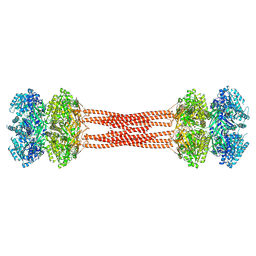 | | Crystal structure of dodecameric KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
7Q51
 
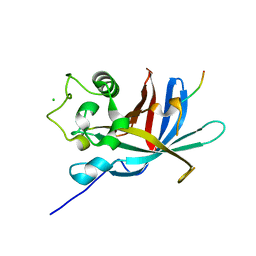 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
1K3E
 
 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
