1HF9
 
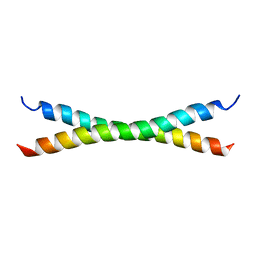 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
7Q1L
 
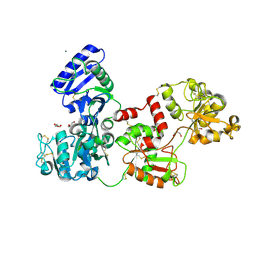 | | Glycosilated Human Serum Apo-tranferrin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Gavira, J.A, Moreno, A, Campos-Escamilla, C, Gonzalez-Ramirez, L.A, Siliqi, D. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin.
Int J Mol Sci, 22, 2021
|
|
6ZWM
 
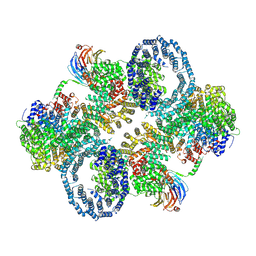 | | cryo-EM structure of human mTOR complex 2, overall refinement | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
4E0O
 
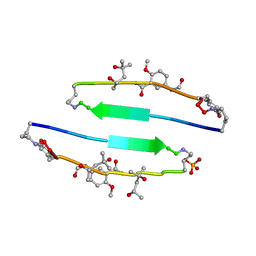 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form III) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7A21
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2158 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type I, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-08-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2153
To Be Published
|
|
7A4U
 
 | |
7PSU
 
 | | Structure of protein kinase CK2alpha mutant K198R associated with the Okur-Chung Neurodevelopmental Syndrome | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Werner, C, Gast, A, Lindenblatt, D, Nickelsen, K, Niefind, K, Jose, J, Hochscherf, J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Enzymological Evidence for an Altered Substrate Specificity in Okur-Chung Neurodevelopmental Syndrome Mutant CK2 alpha Lys198Arg.
Front Mol Biosci, 9, 2022
|
|
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1THD
 
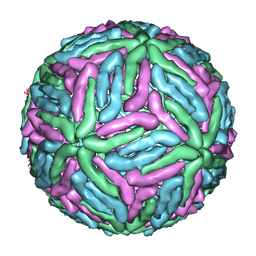 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
7PLM
 
 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
8C77
 
 | | Human cathepsin L after reaction with the thiocarbazate inhibitor CID 16725315 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6ZY1
 
 | |
1HDJ
 
 | | HUMAN HSP40 (HDJ-1), NMR | | Descriptor: | HUMAN HSP40 | | Authors: | Qian, Y.Q, Patel, D, Hartl, F.-U, Mccoll, D.J. | | Deposit date: | 1996-05-09 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain.
J.Mol.Biol., 260, 1996
|
|
1YS5
 
 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
8BZV
 
 | |
1TJE
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase | | Descriptor: | Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1TJP
 
 | | Crystal Structure Of Wild-Type Tryptophan Synthase Complexed With 1-[(2-hydroxylphenyl)amino]3-glycerolphosphate | | Descriptor: | 1-[(2-HYDROXYLPHENYL)AMINO]3-GLYCEROLPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
6ZQI
 
 | | Cryo-EM structure of Spondweni virus prME | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
1T5N
 
 | | Structural transitions as determinants of calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
4HFR
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with an orally bioavailable acidic inhibitor AZD4017. | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {(3S)-1-[5-(cyclohexylcarbamoyl)-6-(propylsulfanyl)pyridin-2-yl]piperidin-3-yl}acetic acid | | Authors: | Ogg, D.J, Gerhardt, S, Hargreaves, D. | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Bioavailable Acidic 11 -Hydroxysteroid Dehydrogenase Type 1 (11 -HSD1) Inhibitor: Discovery of 2-[(3S)-1-[5-(Cyclohexylcarbamoyl)-6-propylsulfanylpyridin-2-yl]-3-piperidyl]acetic Acid (AZD4017)
J.Med.Chem., 55, 2012
|
|
1YO5
 
 | | Analysis of the 2.0A crystal structure of the protein-DNA complex of human PDEF Ets domain bound to the prostate specific antigen regulatory site | | Descriptor: | Enhancer site of Prostate Specific Antigen Promoter Region, SAM pointed domain containing ets transcription factor | | Authors: | Wang, Y, Feng, L, Said, M, Balderman, S, Fayazi, Z, Liu, Y, Ghosh, D, Gulick, A.M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the 2.0 A Crystal Structure of the Protein-DNA Complex of the Human PDEF Ets Domain Bound to the Prostate Specific Antigen Regulatory Site
Biochemistry, 44, 2005
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
1YU7
 
 | | Crystal Structure of the W64Y mutant of Villin Headpiece | | Descriptor: | Villin | | Authors: | Meng, J, Vardar, D, Wang, Y, Guo, H.C, Head, J.F, McKnight, C.J. | | Deposit date: | 2005-02-12 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity.
Biochemistry, 44, 2005
|
|
