2WV3
 
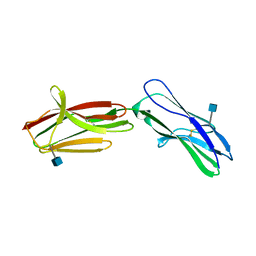 | | Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPLASTIN | | Authors: | Soroka, V, Owczarek, S, Kastrup, J.S, Berezin, V, Bock, E, Gajhede, M. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuroplastin-55 Binds to and Signals Through the Fibroblast Growth Factor Receptor
Faseb J., 24, 2010
|
|
1HZS
 
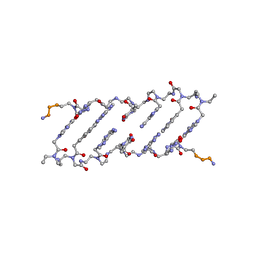 | | Crystal structure of a peptide nucleic acid duplex (BT-PNA) containing a bicyclic analogue of thymine | | Descriptor: | PEPTIDE NUCLEIC ACID | | Authors: | Eldrup, A.B, Nielsen, B.B, Haaima, G, Rasmussen, H, Kastrup, J.S, Christensen, C, Nielsen, P.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,8-Naphthyridin-2(1H)-ones. Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs)
Eur.J.Org.Chem., 9, 2001
|
|
3S2V
 
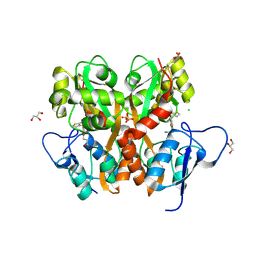 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
1RRU
 
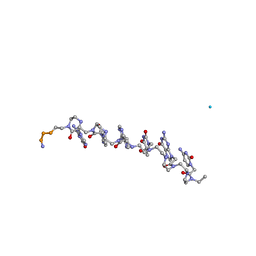 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | Descriptor: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | Authors: | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
1HTN
 
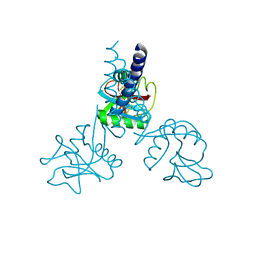 | | HUMAN TETRANECTIN, A TRIMERIC PLASMINOGEN BINDING PROTEIN WITH AN ALPHA-HELICAL COILED COIL | | Descriptor: | CALCIUM ION, TETRANECTIN | | Authors: | Nielsen, B.B, Kastrup, J.S, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thogersen, H.C, Larsen, I.K. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of tetranectin, a trimeric plasminogen-binding protein with an alpha-helical coiled coil.
FEBS Lett., 412, 1997
|
|
4ISU
 
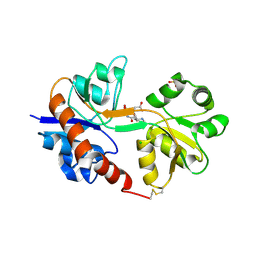 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
4H11
 
 | | Interaction partners of PSD-93 studied by X-ray crystallography and fluorescent polarization spectroscopy | | Descriptor: | ACETATE ION, Disks large homolog 2, SULFATE ION | | Authors: | Fiorentini, M, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Interaction partners of PSD-93 studied by X-ray crystallography and fluorescence polarization spectroscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IGT
 
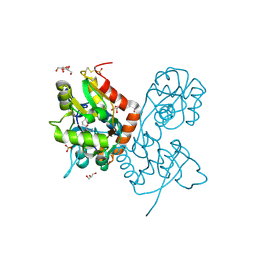 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
4KFQ
 
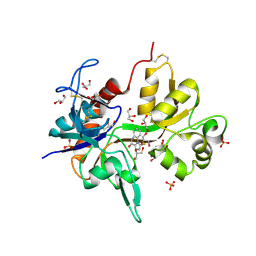 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
2VRK
 
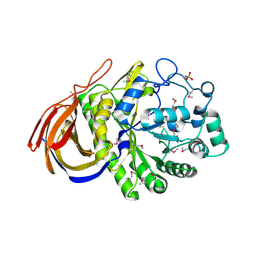 | | Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION | | Authors: | Paes, G, Skov, L.K, ODonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
2WKY
 
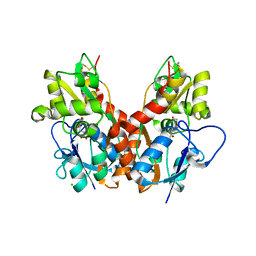 | | Crystal structure of the ligand-binding core of GluR5 in complex with the agonist 4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Glutamate Receptor Glur5 Agonist (S)-2-Amino-3-(3-Hydroxy-7,8-Dihydro-6H-Cyclohepta[D]Isoxazol-4-Yl)Propionic Acid and the 8-Methyl Analogue: Synthesis, Molecular Pharmacology, and Biostructural Characterization
J.Med.Chem., 52, 2009
|
|
2VRQ
 
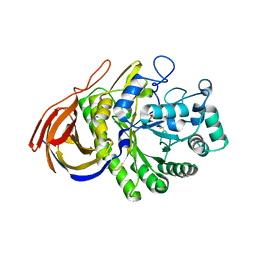 | | STRUCTURE OF AN INACTIVE MUTANT OF ARABINOFURANOSIDASE FROM THERMOBACILLUS XYLANILYTICUS IN COMPLEX WITH A PENTASACCHARIDE | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Paes, G, Skov, L.K, Odonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
2V3T
 
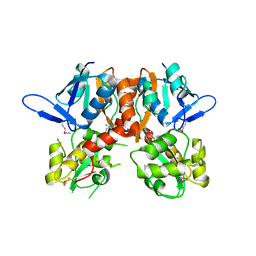 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | Descriptor: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2WL7
 
 | | Crystal structure of the PSD93 PDZ1 domain | | Descriptor: | CHLORIDE ION, DISKS LARGE HOMOLOG 2, SULFATE ION | | Authors: | Fiorentini, M, Kallehauge, A, Kristensen, O, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of the First Pdz Domain of Human Psd-93.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2WPG
 
 | | Sucrose Hydrolase | | Descriptor: | AMYLOSUCRASE OR ALPHA AMYLASE | | Authors: | Champion, E, Remaud-Simeon, M, Skov, L.K, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2009-08-06 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Apo Structure of Sucrose Hydrolase from Xanthomonas Campestris Pv. Campestris Shows an Open Active-Site Groove
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | Descriptor: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2VAJ
 
 | | Crystal structure of NCAM2 Ig1 (I4122 cell unit) | | Descriptor: | NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Kastrup, J.S, Navarro-Poulsen, J.-C, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2007-08-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
J.Mol.Biol., 382, 2008
|
|
6YK4
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound ( S) - 1- [2'-Amino-2'-carboxyethyl]-6-methyl-5 ,7- dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 1.00A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
1A7S
 
 | | ATOMIC RESOLUTION STRUCTURE OF HBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Karlsen, S, Iversen, L.F, Larsen, I.K, Flodgaard, H.J, Kastrup, J.S. | | Deposit date: | 1998-03-17 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of human HBP/CAP37/azurocidin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2P2A
 
 | | X-ray structure of the GluR2 ligand binding core (S1S2J) in complex with 2-Bn-tet-AMPA at 2.26A resolution | | Descriptor: | 2-AMINO-3-[3-HYDROXY-5-(2-BENZYL-2H-5-TETRAZOLYL)-4-ISOXAZOLYL]-PROPIONIC ACID, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A tetrazolyl-substituted subtype-selective AMPA receptor agonist.
J.Med.Chem., 50, 2007
|
|
2FPE
 
 | | Conserved dimerization of the ib1 src-homology 3 domain | | Descriptor: | C-jun-amino-terminal kinase interacting protein 1, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guenat, S, Dar, I, Bonny, C, Kastrup, J.S, Gajhede, M, Kristensen, O. | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique set of SH3-SH3 interactions controls IB1 homodimerization
Embo J., 25, 2006
|
|
5FHO
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-chlorobenzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at 2.3 A resolution | | Descriptor: | (1S)-1-carboxy-2-(5-{2-[(3-chlorophenyl)methyl]-2H-tetrazol-5-yl}-3-oxo-2,3-dihydro-1,2-oxazol-4-yl)ethan-1-aminium, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
5FHN
 
 | |
3TDJ
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-97 at 1.95 A resolution | | Descriptor: | 4-ethyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Krintel, C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-08-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2.
Biochem.J., 441, 2012
|
|
3U8J
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
