6YA9
 
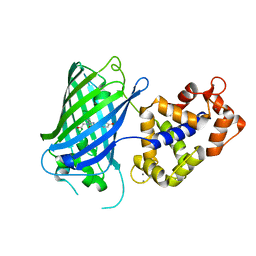 | | Crystal structure of rsGCaMP in the ON state (non-illuminated) | | Descriptor: | CALCIUM ION, rsCGaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-03-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
1FIG
 
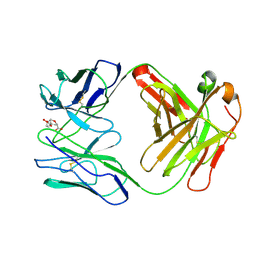 | | ROUTES TO CATALYSIS: STRUCTURE OF A CATALYTIC ANTIBODY AND COMPARISON WITH ITS NATURAL COUNTERPART | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, IGG1-KAPPA 1F7 FAB (HEAVY CHAIN), IGG1-KAPPA 1F7 FAB (LIGHT CHAIN) | | Authors: | Haynes, M.R, Stura, E.A, Hilvert, D, Wilson, I.A. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart.
Science, 263, 1994
|
|
1ZMY
 
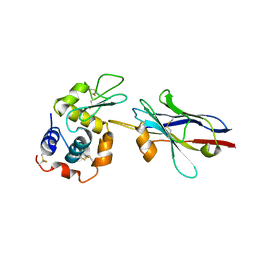 | | cAbBCII-10 VHH framework with CDR loops of cAbLys3 grafted on it and in complex with hen egg white lysozyme | | Descriptor: | Antibody cabbcII-10:lys3, Lysozyme C | | Authors: | Saerens, D, Pellis, M, Loris, R, Pardon, E, Dumoulin, M, Matagne, A, Wyns, L, Muyldermans, S, Conrath, K. | | Deposit date: | 2005-05-11 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies
J.Mol.Biol., 352, 2005
|
|
1GJZ
 
 | |
7P5W
 
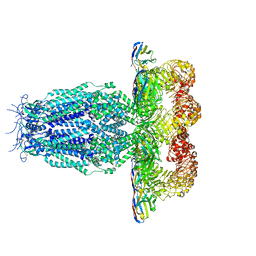 | |
8ALU
 
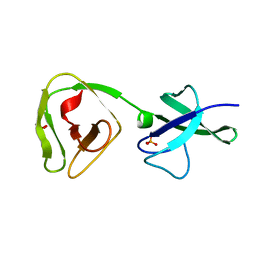 | | Crystal structure of the teichoic acid binding domain of SlpA, S-layer protein from Lactobacillus acidophilus (aa. 314-444) | | Descriptor: | PHOSPHATE ION, S-layer protein | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7P6K
 
 | |
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
3FPW
 
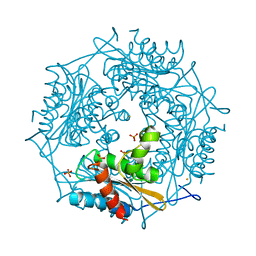 | | Crystal Structure of HbpS with bound iron | | Descriptor: | Extracellular haem-binding protein, FE (III) ION, PHOSPHATE ION | | Authors: | Ortiz de Orue Lucana, D, Bogel, G, Zou, P, Groves, M.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase
J.Mol.Biol., 386, 2009
|
|
7P5Y
 
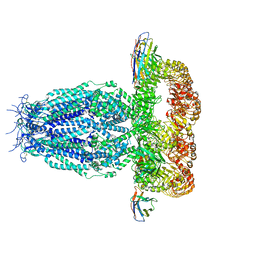 | |
7P5V
 
 | |
1Z5S
 
 | | Crystal structure of a complex between UBC9, SUMO-1, RANGAP1 and NUP358/RANBP2 | | Descriptor: | Ran GTPase-activating protein 1, Ran-binding protein 2, Ubiquitin-conjugating enzyme E2 I, ... | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2005-03-19 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex.
Nature, 435, 2005
|
|
6YDH
 
 | |
7P34
 
 | |
1FZX
 
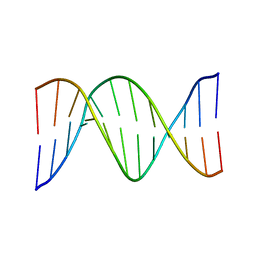 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
6YU6
 
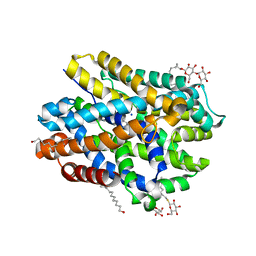 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
1G0F
 
 | |
7P60
 
 | |
3T59
 
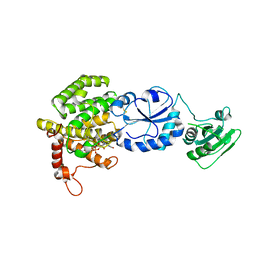 | |
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1TXI
 
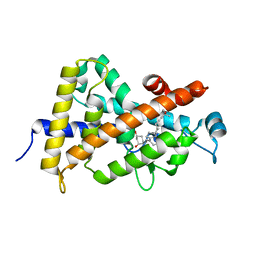 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | Descriptor: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | Authors: | Moras, D, Rochel, N. | | Deposit date: | 2004-07-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6YYI
 
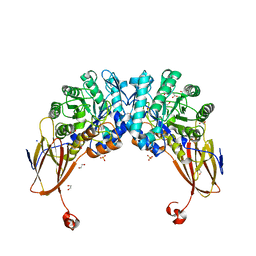 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
6YZ0
 
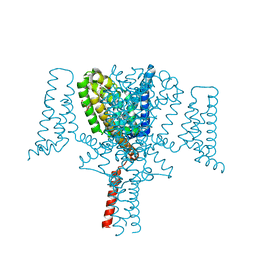 | | Full length Open-form Sodium Channel NavMs F208L in complex with Cannabidiol (CBD) | | Descriptor: | 2-[(1R,2R,5S)-5-methyl-2-(prop-1-en-2-yl)cyclohexyl]-5-pentylbenzene-1,3-diol, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Sait, L.G, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cannabidiol interactions with voltage-gated sodium channels.
Elife, 9, 2020
|
|
1Z9W
 
 | | Tetrameric structure of apo-7,8-Dihydroneopterin Aldolase from Mycobacterium tuberculosis | | Descriptor: | Dihydroneopterin aldolase | | Authors: | Goulding, C.W, Apostol, M.I, Sawaya, M.R, Philips, M, Parseghian, A, Eisenberg, D, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-04-05 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme.
J.Mol.Biol., 349, 2005
|
|
