3CXY
 
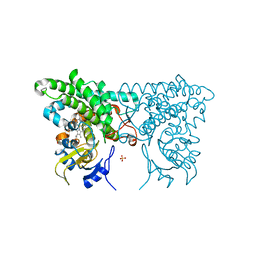 | |
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
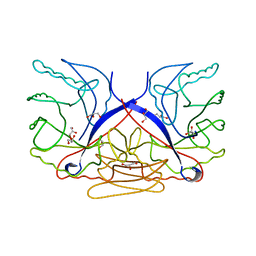
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
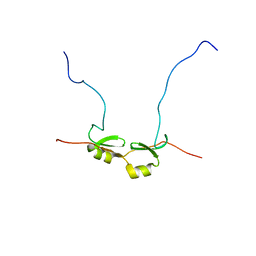
1SEQ
 
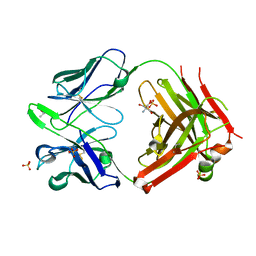 | | Fab MNAC13 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, Monoclonal Antibody MNAC13, ... | | Authors: | Covaceuszach, S, Cattaneo, A, Lamba, D. | | Deposit date: | 2004-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Neutralization of NGF-TrkA Receptor Interaction by the Novel Antagonistic anti-TrkA Monoclonal Antibody MNAC13: a Structural Insight
Proteins, 58, 2005
|
|
3AK4
 
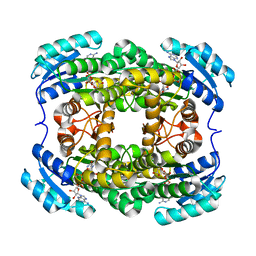 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
2MN5
 
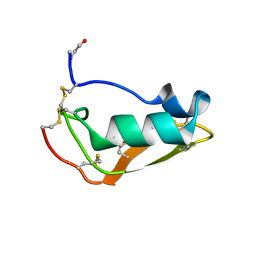 | | NMR structure of Copsin | | Descriptor: | Copsin | | Authors: | Hofmann, D, Wider, G, Essig, A, Aebi, M. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Copsin, a Novel Peptide-based Fungal Antibiotic Interfering with the Peptidoglycan Synthesis.
J.Biol.Chem., 289, 2014
|
|
2DWM
 
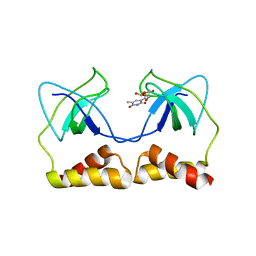 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
1SJ5
 
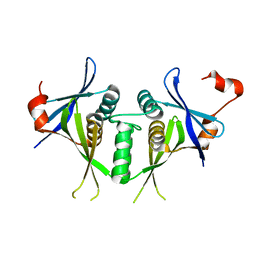 | | Crystal structure of a duf151 family protein (tm0160) from thermotoga maritima at 2.8 A resolution | | Descriptor: | conserved hypothetical protein TM0160 | | Authors: | Spraggon, G, Panatazatos, D, Klock, H.E, Wilson, I.A, Woods Jr, V.L, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-03-02 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM1171.
Protein Sci., 13, 2004
|
|
372D
 
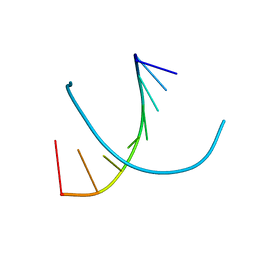 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
1NAN
 
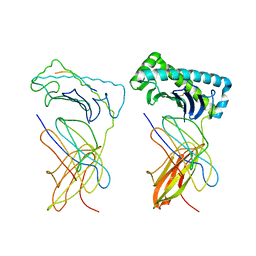 | | MCH CLASS I H-2KB MOLECULE COMPLEXED WITH PBM1 PEPTIDE | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
3A40
 
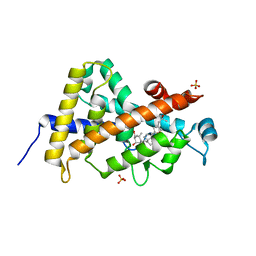 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277B(C23R) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-2-methyl-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
3A4L
 
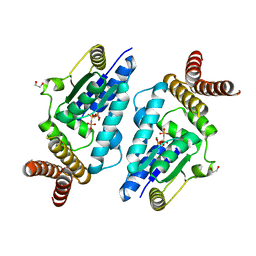 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AAV
 
 | | Bovine beta-trypsin bound to meta-diamidino schiff base copper (II) chelate | | Descriptor: | 3,3'-[ethane-1,2-diylbis(nitrilomethylylidene)]bis(4-hydroxybenzenecarboximidamide), CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
2GG1
 
 | | NMR solution structure of domain III of the E-protein of tick-borne Langat flavivirus (includes RDC restraints) | | Descriptor: | Genome polyprotein | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
1IKF
 
 | | A CONFORMATION OF CYCLOSPORIN A IN AQUEOUS ENVIRONMENT REVEALED BY THE X-RAY STRUCTURE OF A CYCLOSPORIN-FAB COMPLEX | | Descriptor: | CYCLOSPORIN A, IGG1-KAPPA R45-45-11 FAB (HEAVY CHAIN), IGG1-KAPPA R45-45-11 FAB (LIGHT CHAIN) | | Authors: | Vix, O, Altschuh, D, Rees, B, Thierry, J.-C. | | Deposit date: | 1993-12-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformation of Cyclosporin a in Aqueous Environment Revealed by the X-Ray Structure of a Cyclosporin-Fab Complex.
Science, 256, 1992
|
|
2FQH
 
 | | NMR structure of hypothetical protein TA0938 from Termoplasma acidophilum | | Descriptor: | Hypothetical protein TA0938 | | Authors: | Monleon, D, Esteve, V, Yee, A, Arrowsmith, C, Celda, B, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of hypothetical protein TA0938 from Thermoplasma acidophilum.
Proteins, 67, 2007
|
|
2FO8
 
 | | Solution structure of the Trypanosoma cruzi cysteine protease inhibitor chagasin | | Descriptor: | Chagasin | | Authors: | Salmon, D, do Aido-Machado, R, de Lima, A.A.P, Scharfstein, J, Oschkinat, H, Pires, J.R. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Trypanosoma cruzi Cysteine Protease Inhibitor Chagasin
J.Mol.Biol., 357, 2006
|
|
3AEL
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing methionine imine-pyridoxamine-5'-phosphate and alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AGP
 
 | | Structure of viral polymerase form I | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2GSV
 
 | | X-Ray Crystal Structure of Protein YvfG from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR478. | | Descriptor: | Hypothetical protein yvfG, SULFATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Wang, D, Fang, Y, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Hypothetical Protein YvfG from Bacillus subtilis, Northeast Structural Genomics Target SR478
To be Published
|
|
2GPT
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase/ shikimate 5-dehydrogenase, L(+)-TARTARIC ACID, ... | | Authors: | Singh, S.A, Christendat, D. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway
Biochemistry, 45, 2006
|
|
2GU1
 
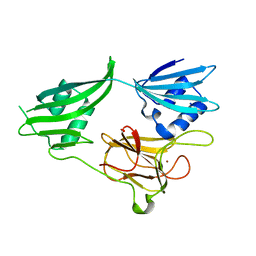 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
3CW4
 
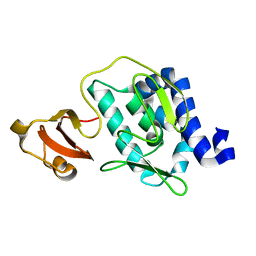 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
1JBU
 
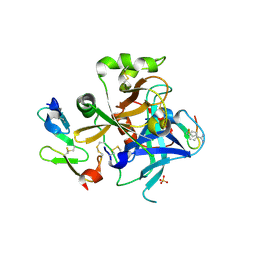 | | Coagulation Factor VII Zymogen (EGF2/Protease) in Complex with Inhibitory Exosite Peptide A-183 | | Descriptor: | BENZAMIDINE, COAGULATION FACTOR VII, Peptide exosite inhibitor A-183, ... | | Authors: | Eigenbrot, C, Kirchhofer, D, Dennis, M.S, Santell, L, Lazarus, R.A, Stamos, J, Ultsch, M.H. | | Deposit date: | 2001-06-06 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The factor VII zymogen structure reveals reregistration of beta strands during activation.
Structure, 9, 2001
|
|
2GTH
 
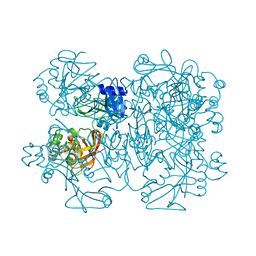 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
