2REN
 
 | | STRUCTURE OF RECOMBINANT HUMAN RENIN, A TARGET FOR CARDIOVASCULAR-ACTIVE DRUGS, AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RENIN | | Authors: | Sielecki, A.R, James, M.N.G. | | Deposit date: | 1992-02-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 A resolution.
Science, 243, 1989
|
|
2RDL
 
 | | Hamster Chymase 2 | | Descriptor: | Chymase 2, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION | | Authors: | Spurlino, J, Abad, M, Kervinen, J. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-30 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for elastolytic substrate specificity in rodent alpha-chymases.
J.Biol.Chem., 283, 2008
|
|
1O6B
 
 | |
1O63
 
 | |
1O6C
 
 | |
2RF7
 
 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
2RG5
 
 | | Phenylalanine pyrrolotriazine p38 alpha map kinase inhibitor compound 11B | | Descriptor: | Mitogen-activated protein kinase 14, N-ethyl-4-{[5-(methoxycarbamoyl)-2-methylphenyl]amino}-5-methylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide | | Authors: | Sack, J.S. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Synthesis, and Anti-inflammatory Properties of Orally Active 4-(Phenylamino)-pyrrolo[2,1-f][1,2,4]triazine p38alpha Mitogen-Activated Protein Kinase Inhibitors
J.Med.Chem., 51, 2008
|
|
1O66
 
 | |
1O60
 
 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O67
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
3V6Q
 
 | | Crystal structure of the complex of bovine lactoperoxidase with Carbon monoxide at 2.0 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bovine carbonyl lactoperoxidase structure at 2.0 angstrom resolution and infrared spectra as a function of pH.
Protein J., 31, 2012
|
|
3UW4
 
 | | Crystal structure of cIAP1 BIR3 bound to GDC0152 | | Descriptor: | Baculoviral IAP repeat-containing protein 2, Baculoviral IAP repeat-containing protein 4, GDC0152, ... | | Authors: | Maurer, B, Hymowitz, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152).
J.Med.Chem., 55, 2012
|
|
3UW5
 
 | | Crystal structure of the BIR domain of MLIAP bound to GDC0152 | | Descriptor: | Baculoviral IAP repeat-containing protein 7, Baculoviral IAP repeat-containing protein 4, GDC-0152, ... | | Authors: | Maurer, B, Hymowitz, S.G. | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152).
J.Med.Chem., 55, 2012
|
|
3V6R
 
 | |
3V6S
 
 | |
3V7I
 
 | |
3USN
 
 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THE THIADIAZOLE INHIBITOR IPNU-107859, NMR, 1 STRUCTURE | | Descriptor: | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2-YL)-UREIDO]-N-METHYL-3-PHENYL-PROPIONAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Stockman, B.J. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of stromelysin complexed to thiadiazole inhibitors.
Protein Sci., 7, 1998
|
|
3VEV
 
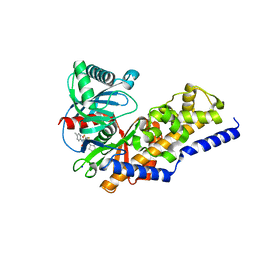 | | Glucokinase in complex with an activator and glucose | | Descriptor: | (2S)-3-cyclopentyl-N-(5-methylpyridin-2-yl)-2-[2-oxo-4-(trifluoromethyl)pyridin-1(2H)-yl]propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VF6
 
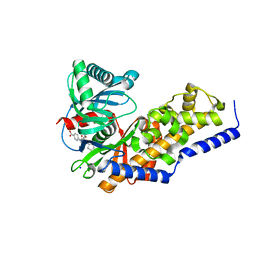 | | Glucokinase in complex with glucose and activator | | Descriptor: | 6-({(2S)-3-cyclopentyl-2-[4-(trifluoromethyl)-1H-imidazol-1-yl]propanoyl}amino)pyridine-3-carboxylic acid, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VZL
 
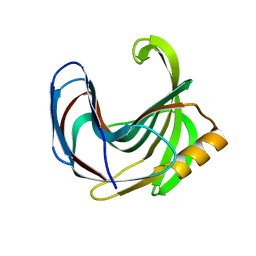 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
7ASS
 
 | |
7AEL
 
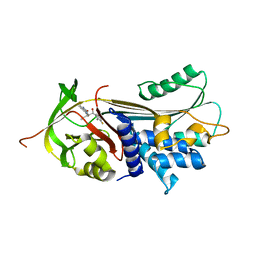 | | alpha 1-antitrypsin (C232S) complexed with GSK716 | | Descriptor: | Alpha-1-antitrypsin, SULFATE ION, ~{N}-[(1~{S},2~{R})-1-(3-fluoranyl-2-methyl-phenyl)-1-oxidanyl-pentan-2-yl]-2-oxidanylidene-1,3-dihydroindole-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of a small molecule that corrects misfolding and increases secretion of Z alpha 1 -antitrypsin.
Embo Mol Med, 13, 2021
|
|
7AK9
 
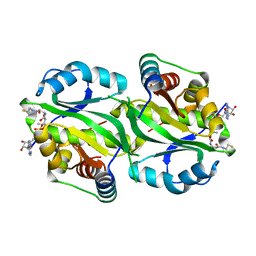 | | Structure of Salmonella TacT3 toxin bound to TacA3 antitoxin C-terminal peptide | | Descriptor: | ABC transporter, Acetyltransferase, DEPHOSPHO COENZYME A | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK7
 
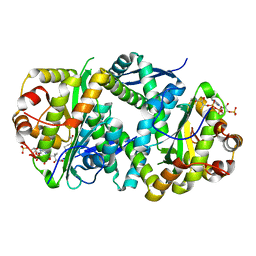 | | Structure of Salmonella TacT2 toxin bound to TacA2 antitoxin | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CHLORIDE ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK8
 
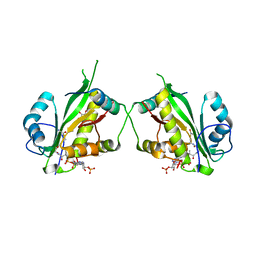 | | Structure of Salmonella TacT1 toxin bound to TacA1 antitoxin C-terminal peptide | | Descriptor: | ACETYL COENZYME *A, GCN5 family acetyltransferase, GLYCEROL, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S, Hare, S.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
