3VS3
 
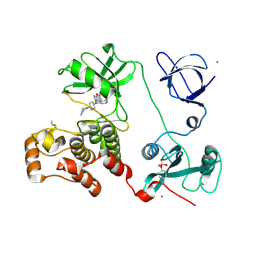 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS0
 
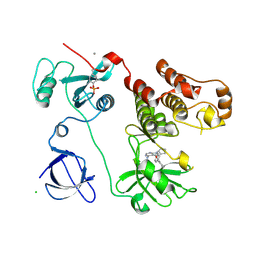 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
2DCE
 
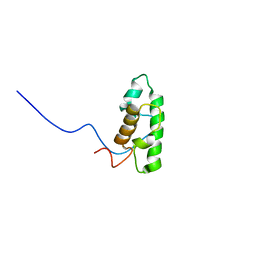 | | Solution structure of the SWIRM domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
3VS5
 
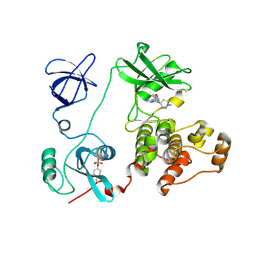 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
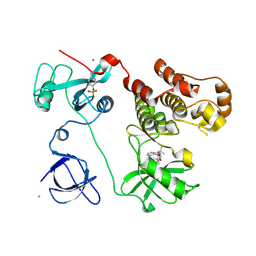 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3KC6
 
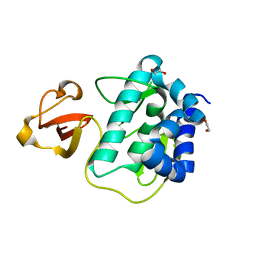 | |
7CZF
 
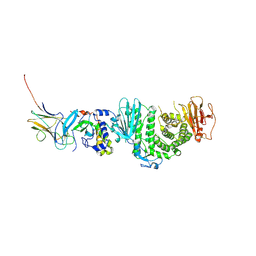 | | Crystal structure of Kaposi Sarcoma associated herpesvirus (KSHV ) gHgL in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
3KHW
 
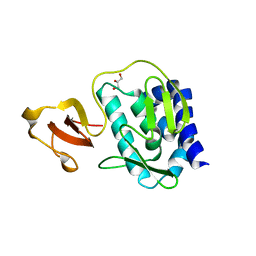 | |
7CZE
 
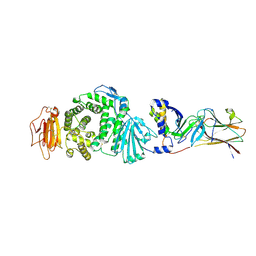 | | Crystal structure of Epstein-Barr virus (EBV) gHgL and in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
2CU7
 
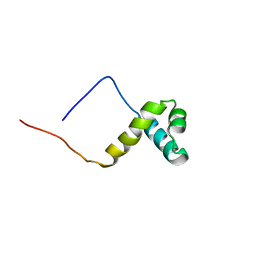 | | Solution structure of the SANT domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Umehara, T, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-25 | | Release date: | 2005-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2COM
 
 | | The solution structure of the SWIRM domain of human LSD1 | | Descriptor: | Lysine-specific histone demethylase 1 | | Authors: | Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-18 | | Release date: | 2005-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIRM domain of human histone demethylase LSD1
Structure, 14, 2006
|
|
3B3O
 
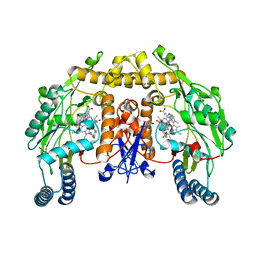 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
2D8B
 
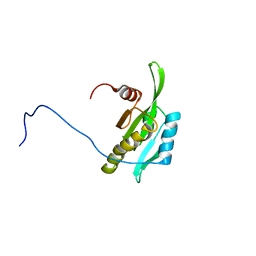 | | Solution structure of the second tandem cofilin-domain of mouse twinfilin | | Descriptor: | Twinfilin-1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains
Protein Sci., 18, 2009
|
|
6JLH
 
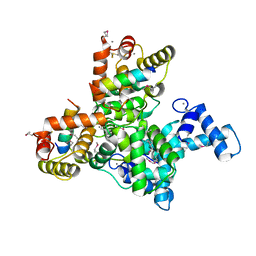 | | Structure of SCGN in complex with a Snap25 peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secretagogin, ... | | Authors: | Qin, J, Sun, Q, Jia, D. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and mechanistic insights into secretagogin-mediated exocytosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2E63
 
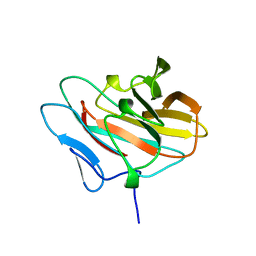 | | Solution structure of the NEUZ domain in KIAA1787 protein | | Descriptor: | KIAA1787 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
2CXF
 
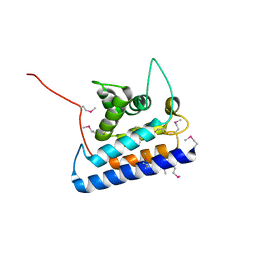 | | RUN domain of Rap2 interacting protein x, crystallized in C2 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWG
 
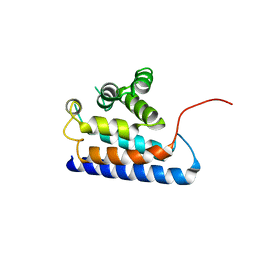 | | RUN domain of Rap2 interacting protein x, crystallized in P2(1)2(1)2(1) space group | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2E5S
 
 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
8BFW
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-27 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8BN1
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-4PH-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-11-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8B9P
 
 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8BYJ
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
2CXL
 
 | | RUN domain of Rap2 interacting protein x, crystallized in I422 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Umehara, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWK
 
 | | Crystal structure of the RUN domain of mouse Rap2 interacting protein x | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2YTX
 
 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
