8HDD
 
 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
6M9L
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6M95
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
5Z0U
 
 | |
2D5K
 
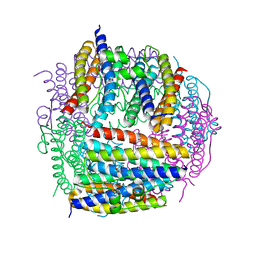 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
5Z0T
 
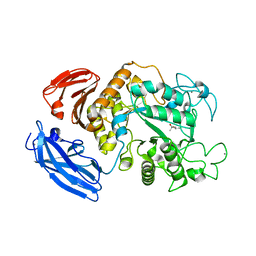 | |
6JU1
 
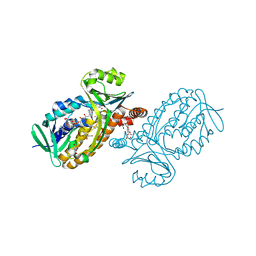 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
2ZP0
 
 | | Human factor viia-tissue factor complexed with benzylsulfonamide-D-ile-gln-P-aminobenzamidine | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-2-[[(2R,3R)-3-methyl-2-(phenylmethylsulfonylamino)pentanoyl]amino]pentanediamide, CALCIUM ION, Factor VII heavy chain, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Mimetic Factor VIIa Inhibitor: Importance of Hydrophilic Pocket in S2 Site to Improve Selectivity aganist Thrombin
LETT.DRUG DES.DISCOVERY, 2, 2005
|
|
7DDW
 
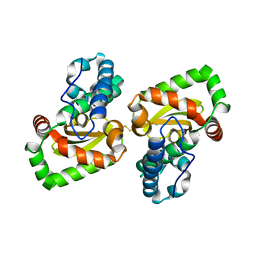 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase S126C | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-10-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The role of S126 in the Staphylococcus equorum MnSOD activity and stability.
J.Struct.Biol., 213, 2021
|
|
6M30
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase N73F | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unprecedented Role of The N73-F124 Pair in The Staphylococcus equorum MnSOD Activity.
Curr Enzym Inhib, 2021
|
|
6J6Y
 
 | | FGFR4 D2 - Fab complex | | Descriptor: | Fab Heavy chain, Fab light chain, Fibroblast growth factor receptor 4 | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Preclinical Development of U3-1784, a Novel FGFR4 Antibody Against Cancer, and Avoidance of Its On-target Toxicity.
Mol.Cancer Ther., 18, 2019
|
|
1FIA
 
 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
4JMU
 
 | | Crystal structure of HIV matrix residues 1-111 in complex with inhibitor | | Descriptor: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, Gag-Pol polyprotein, SULFATE ION | | Authors: | Lemke, C.T. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
4J93
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
4JVQ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 9 | | Descriptor: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-26 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
4INB
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With benzodiazepine Inhibitor | | Descriptor: | (3Z)-3-{[(2-methoxyethyl)amino]methylidene}-1-methyl-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, Gag protein, SODIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monitoring Binding of HIV-1 Capsid Assembly Inhibitors Using (19) F Ligand-and (15) N Protein-Based NMR and X-ray Crystallography: Early Hit Validation of a Benzodiazepine Series.
Chemmedchem, 8, 2013
|
|
2E2B
 
 | | Crystal structure of the c-Abl kinase domain in complex with INNO-406 | | Descriptor: | N-[3-(4,5'-BIPYRIMIDIN-2-YLAMINO)-4-METHYLPHENYL]-4-{[(3S)-3-(DIMETHYLAMINO)PYRROLIDIN-1-YL]METHYL}-3-(TRIFLUOROMETHYL) BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Horio, T, Hamasaki, T, Wakayama, T, Takagaki, K, Ohgi, T. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural factors contributing to the Abl/Lyn dual inhibitory activity of 3-substituted benzamide derivatives
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3WR7
 
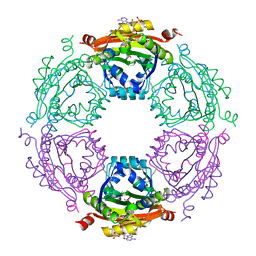 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
4DWW
 
 | |
7Z3W
 
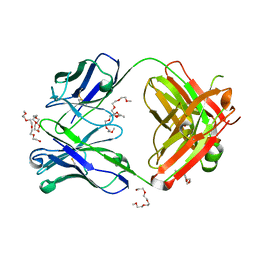 | | Crystal structure of the AAL160 Fab | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AAL160 Fab heavy-chain, AAL160 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z4T
 
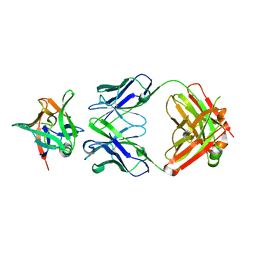 | | AAL160 FAB IN COMPLEX WITH HUMAN INTERLEUKIN-1 BETA | | Descriptor: | AAL160 Fab heavy-chain, AAL160 light-chain, Interleukin-1 beta | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z2M
 
 | |
1R6K
 
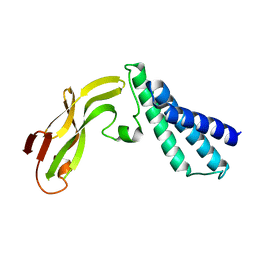 | | HPV11 E2 TAD crystal structure | | Descriptor: | HPV11 REGULATORY PROTEIN E2 | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1R6N
 
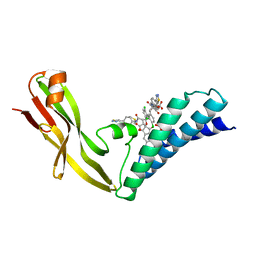 | | HPV11 E2 TAD complex crystal structure | | Descriptor: | 2-METHYL-PROPIONIC ACID, DIMETHYL SULFOXIDE, HPV11 REGULATORY PROTEIN E2, ... | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human
Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
3AJJ
 
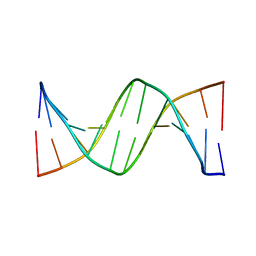 | | Crystal Structure of d(CGCGGATf5UCGCG): 5-Formyluridine/Guanosine Base-pair in B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
