1X94
 
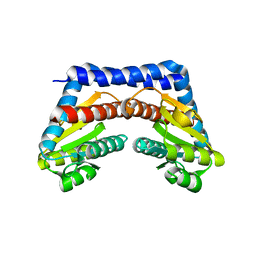 | |
1ZL6
 
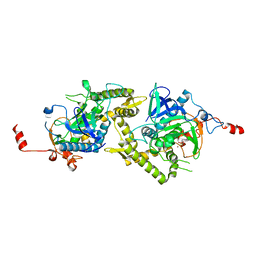 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZN3
 
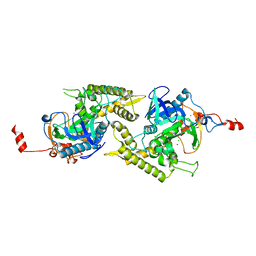 | | Crystal structure of Glu335Ala mutant of Clostridium botulinum neurotoxin type E | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1SKU
 
 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
1TGU
 
 | | The crystal structure of bovine liver catalase without NADPH | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-05-31 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
1TXN
 
 | |
1U8S
 
 | |
1U05
 
 | |
1TY8
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
To be Published
|
|
7DAM
 
 | |
7DAH
 
 | |
1TH4
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with 3-amino-1,2,4-triazole | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1TH2
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with azide | | Descriptor: | AZIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1SRU
 
 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1TH3
 
 | | Crystal structure of NADPH depleted bovine live catalase complexed with cyanide | | Descriptor: | CYANIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1SGM
 
 | |
3T2M
 
 | | Crystal Structure of NaK Channel N68D Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T1C
 
 | | Crystal Structure of NaK Channel D66Y Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QW5
 
 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor RRGF | | Descriptor: | Botulinum neurotoxin type A, SULFATE ION, ZINC ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3QW8
 
 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor CRGC | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, SODIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RYS
 
 | |
3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
3TCU
 
 | | Crystal Structure of NaK2K Channel D68E Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
