3STA
 
 | | Crystal structure of ClpP in tetradecameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
5TUB
 
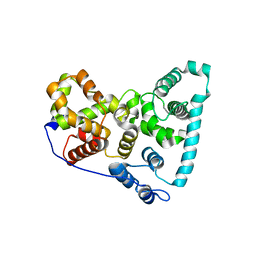 | | Crystal Structure of the Shark TBC1D15 GAP Domain | | Descriptor: | Shark TBC1D15 GTPase-activating Protein | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TUC
 
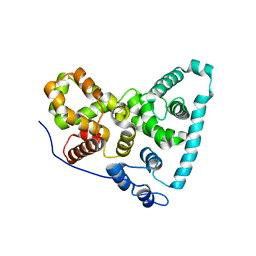 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | Descriptor: | Sus TBC1D15 GAP Domain | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
4X6R
 
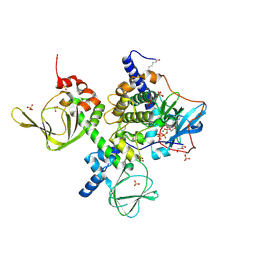 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Wu, J, Taylor, S.S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-07-22 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4X6Q
 
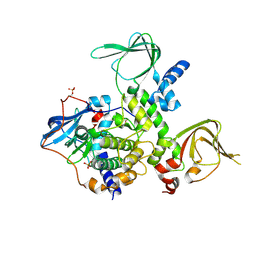 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Taylor, S.S. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
2NA8
 
 | | Transmembrane Structure of the Cytokine Receptor Common Subunit beta | | Descriptor: | Cytokine receptor common subunit beta | | Authors: | Schmidt, T, Ye, F, Situ, A.J, An, W, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors.
J.Biol.Chem., 291, 2016
|
|
2NA9
 
 | | Transmembrane Structure of the P441A Mutant of the Cytokine Receptor Common Subunit beta | | Descriptor: | Cytokine receptor common subunit beta | | Authors: | Schmidt, T, Ye, F, Situ, A.J, An, W, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors.
J.Biol.Chem., 291, 2016
|
|
4R8G
 
 | | Crystal Structure of Myosin-1c tail in complex with Calmodulin | | Descriptor: | Calmodulin, SULFATE ION, Unconventional myosin-Ic | | Authors: | Lu, Q, Li, J, Ye, F, Zhang, M. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure of myosin-1c tail bound to calmodulin provides insights into calcium-mediated conformational coupling.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6JHY
 
 | |
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6JFW
 
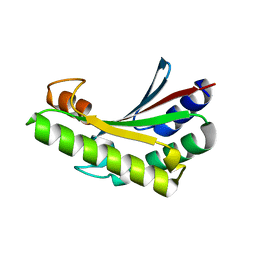 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8JJP
 
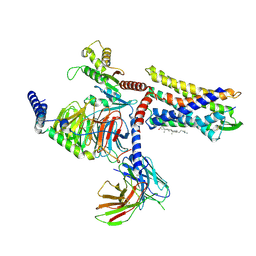 | | G protein-coupled receptor 1 | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, A, Liu, Y, Chen, G, Ye, F. | | Deposit date: | 2023-05-31 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | G protein-coupled receptor 1
To Be Published
|
|
2M9H
 
 | |
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
4EJQ
 
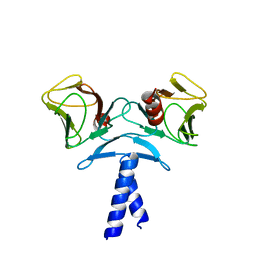 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
7QXI
 
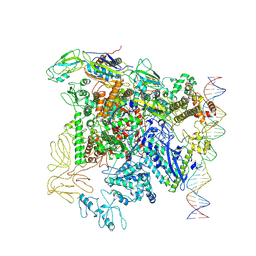 | |
7QV9
 
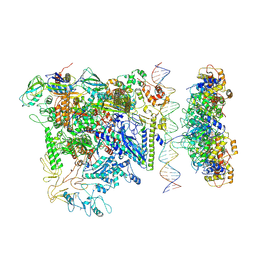 | |
7QWP
 
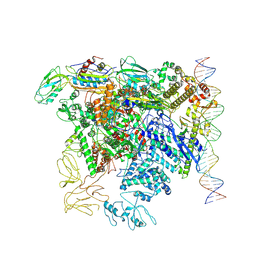 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
