1CVB
 
 | |
1ZSP
 
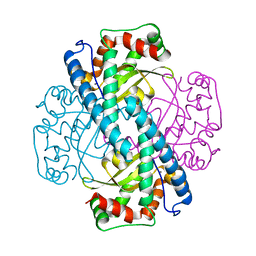 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1ZRB
 
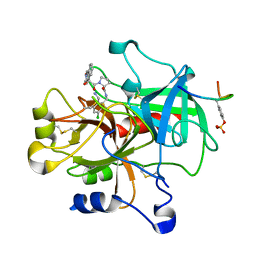 | | Thrombin in complex with an azafluorenyl inhibitor 23b | | Descriptor: | 11-peptide hirudin fragment, 3-AZA-9-HYDROXY-9-FLUORENYLCARBONYL-L-PROLYL-2-AMINOMETHYL-5-CHLOROBENZYLAMIDE, N-OXIDE, ... | | Authors: | Stauffer, K.J, Williams, P.D, Selnick, H.G, Nantermet, P.G, Newton, C.L, Homnick, C.F, Zrada, M.M, Lewis, S.D, Lucas, B.J, Krueger, J.A, Pietrak, B.L, Lyle, E.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Wallace, A.A, Sitko, G.R, Cook, J.J, Holahan, M.A, Stranieri-Michener, M, Leonard, Y.M, Lynch Jr, J.J, McMasters, D.R, Yan, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-07 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 9-hydroxyazafluorenes and their use in thrombin inhibitors
J.Med.Chem., 48, 2005
|
|
4JD7
 
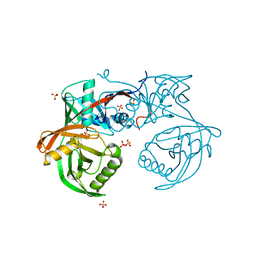 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
4JED
 
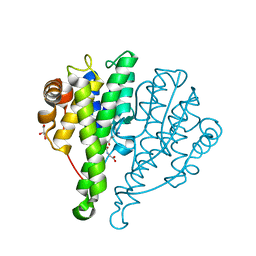 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
1CEQ
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1ZTE
 
 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Suerpoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
4JFY
 
 | | Apo structure of phosphotyrosine (pYAb) scaffold | | Descriptor: | Fab heavy chain, Fab light chain, TETRAETHYLENE GLYCOL | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
4IQ1
 
 | | Crystal structure of glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, SUBSTRATE-FREE | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, AL Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of probable glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, Substrate-free
To be Published
|
|
4IL0
 
 | | Crystal structure of GlucDRP from E. coli K-12 MG1655 (EFI target EFI-506058) | | Descriptor: | CITRIC ACID, GLYCEROL, Glucarate dehydratase-related protein | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-28 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
1YSC
 
 | |
1YO2
 
 | | Proton Transfer from His200 in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Bhatt, D, Tu, C, Fisher, S.Z, Hernandez Prada, J.A, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-01-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton transfer in a Thr200His mutant of human carbonic anhydrase II
Proteins, 61, 2005
|
|
1YU1
 
 | | Major Tropism Determinant P3c Variant | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, Major Tropism Determinant (Mtd-P3c) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
4IJI
 
 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
1YU2
 
 | | Major Tropism Determinant M1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-M1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU0
 
 | | Major Tropism Determinant P1 Variant | | Descriptor: | CALCIUM ION, Major Tropism Determinant (Mtd-P1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU3
 
 | | Major Tropism Determinant I1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-I1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
4IWC
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with a Dynamic Thiophene-derivative | | Descriptor: | 4,4'-thiene-2,5-diylbis(3-methylphenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IY2
 
 | | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4 | | Descriptor: | Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Encinar, J.A, Spiwok, V, Gomez-Garcia, I, Oyenarte, I, Ereno-Orbea, J, Terashima, H, Accardi, A, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To be Published
|
|
4IW9
 
 | | Crystal structure of glutathione s-transferase mha_0454 (target efi-507015) from mannheimia haemolytica, gsh complex | | Descriptor: | ACETATE ION, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Probable Glutathione S-Transferase Mha_0454 (Target Efi-507015) from Mannheimia Haemolytica
To be Published
|
|
152D
 
 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
4IY4
 
 | | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4 | | Descriptor: | Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Encinar, J.A, Spiwok, V, Gomez-Garcia, I, Oyenarte, I, Ereno-Orbea, J, Terashima, H, Accardi, A, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To be Published
|
|
1YW2
 
 | | Mutated Mus Musculus P38 Kinase (mP38) | | Descriptor: | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Laughlin, S.K, Clark, M.P, Djung, J.F, Golebiowski, A, Brugel, T.A, Sabat, M, Bookland, R.G, Laufersweiler, M.J, Vanrens, J.C, Townes, J.A, De, B, Hsieh, L.C, Xu, S.C, Walter, R.L, Mekel, M.J, Janusz, M.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The development of new isoxazolone based inhibitors of tumor necrosis factor-alpha (TNF-alpha) production.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4IZG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | Descriptor: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
