2Y1X
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | CHLORIDE ION, HISTONE-ARGININE METHYLTRANSFERASE CARM1, N-(3-{5-[5-(1H-INDOL-4-YL)-1,3,4-OXADIAZOL-2-YL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL}BENZYL)-L-ALANINAMIDE, ... | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
2XLI
 
 | | Crystal structure of the Csy4-crRNA complex, monoclinic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
3H94
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
2YOF
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- beta-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]thiourea, ACETATE ION, THYMIDYLATE KINASE, ... | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
3H1P
 
 | |
2YBD
 
 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
2YDJ
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-01-25 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
2YNC
 
 | | Plasmodium vivax N-myristoyltransferase in complex with YnC12-CoA thioester. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2YNX
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Archaea Common Ancestor (LACA) from the Precambrian Period | | Descriptor: | ACETATE ION, LACA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
3HBG
 
 | | Structure of recombinant Chicken Liver Sulfite Oxidase mutant C185S | | Descriptor: | HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, Sulfite Oxidase mutant C185S | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
3DFY
 
 | | Crystal structure of apo dipeptide epimerase from Thermotoga maritima | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
3DDP
 
 | |
3DDM
 
 | | CRYSTAL STRUCTURE OF MANDELATE RACEMASE/MUCONATE LACTONIZING ENZYME FROM Bordetella bronchiseptica RB50 | | Descriptor: | Putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-05 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE/MUCONATE
LACTONIZING ENZYME FROM Bordetella bronchiseptica RB50
To be Published
|
|
3DG3
 
 | | Crystal structure of muconate lactonizing enzyme from Mucobacterium Smegmatis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DGB
 
 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DAQ
 
 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
3H5F
 
 | | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides. | | Descriptor: | ACETATE ION, CHLORIDE ION, COIL SER L16L-Pen, ... | | Authors: | Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2009-04-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HCL
 
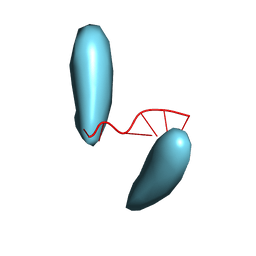 | | Helical superstructures in a DNA oligonucleotide crystal | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | Authors: | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3EAH
 
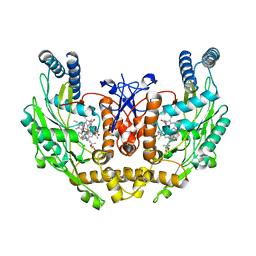 | | Structure of inhibited human eNOS oxygenase domain | | Descriptor: | (3S,5E)-3-propyl-3,4-dihydrothieno[2,3-f][1,4]oxazepin-5(2H)-imine, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3ENE
 
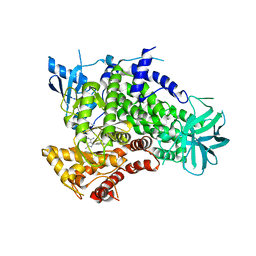 | | Complex of PI3K gamma with an inhibitor | | Descriptor: | 1-methyl-3-naphthalen-2-yl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Apsel, B, Blair, J.A, Gonzalez, B.Z, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases
Nat.Chem.Biol., 4, 2008
|
|
3HBQ
 
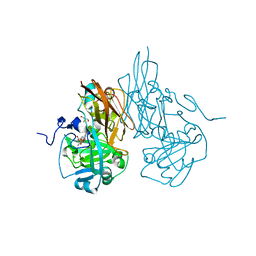 | | Structure of recombinant Chicken Liver Sulfite Oxidase mutant Cys 185 Ala | | Descriptor: | GLYCEROL, HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
3HC2
 
 | | Crystal Structure of chicken sulfite oxidase mutant Tyr 322 Phe | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, sulfite oxidase | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-05 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure function studies in sulfite oxidase with altered active sites
To be Published
|
|
3HGJ
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
