7CI8
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIE
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-3-oxidanyl-N-[3-(trifluoromethyloxy)phenyl]propanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI6
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[3-(4-chlorophenyl)propyl]imidazol-2-yl]ethanol, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIA
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
3AM2
 
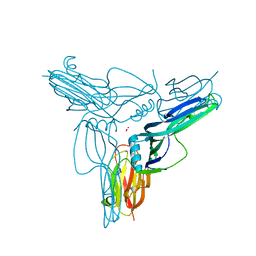 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
5X17
 
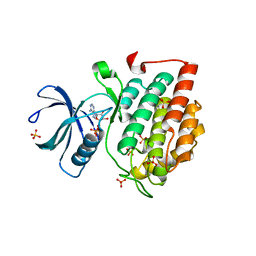 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
7DCF
 
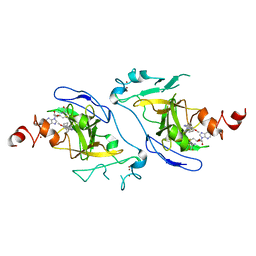 | | Crystal structure of EHMT2 SET domain in complex with compound 10 | | Descriptor: | 5'-methoxy-6'-(1-methyl-2,3,4,7-tetrahydroazepin-5-yl)spiro[cyclobutane-1,3'-indole]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Katayama, K. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS79932728: A Potent, Orally Available G9a/GLP Inhibitor for Treating beta-Thalassemia and Sickle Cell Disease.
Acs Med.Chem.Lett., 12, 2021
|
|
6T9C
 
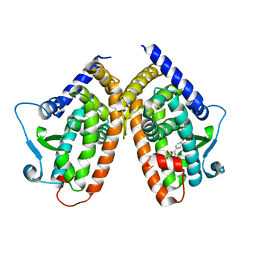 | |
8EEX
 
 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
8EEY
 
 | | Cas7-11 in complex with DR-mismatched target RNA, Csx29 and Csx30 | | Descriptor: | Cas7-11, Csx29, Csx30, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
3ACA
 
 | |
3AC9
 
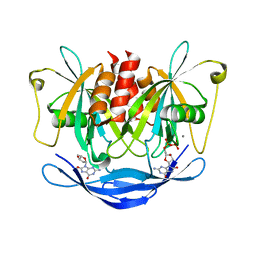 | |
7E7F
 
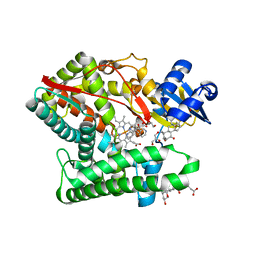 | | Human CYP11B1 mutant in complex with metyrapone | | Descriptor: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
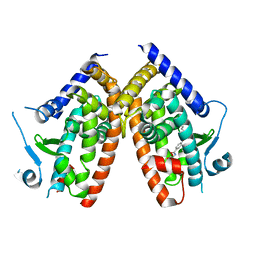
6ZLY
 
 | |
6RS2
 
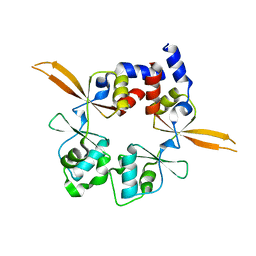 | | Structure of the Bateman module of human CNNM4. | | Descriptor: | Metal transporter CNNM4 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Gomez-Garcia, I, Oyenarte, I, Gimenez, P, Ereno-Orbea, J, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2019-05-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.694 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
3AY5
 
 | |
6G52
 
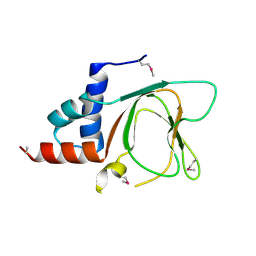 | | CRYSTAL STRUCTURE OF THE CNMP BINDING DOMAIN OF THE MAGNESIUM TRANSPORTER CNNM4 | | Descriptor: | Metal transporter CNNM4 | | Authors: | Gimenez, P, Oyenarte, I, Hardy, S, Zubillaga, M, Merino, N, Blanco, F.J, Siliqi, D, Tremblay, M, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.691 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
1B4U
 
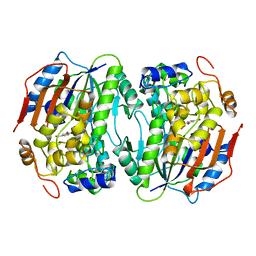 | | PROTOCATECHUATE 4,5-DIOXYGENASE (LIGAB) IN COMPLEX WITH PROTOCATECHUATE (PCA) | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 4,5-DIOXYGENASE | | Authors: | Sugimoto, K, Senda, T, Mitsui, Y. | | Deposit date: | 1998-12-29 | | Release date: | 1999-08-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure Fold.Des., 7, 1999
|
|
1BOU
 
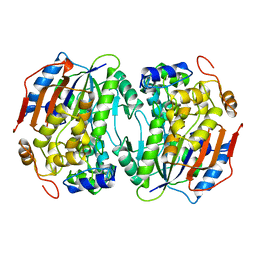 | | THREE-DIMENSIONAL STRUCTURE OF LIGAB | | Descriptor: | 4,5-DIOXYGENASE ALPHA CHAIN, 4,5-DIOXYGENASE BETA CHAIN, FE (III) ION | | Authors: | Sugimoto, K, Senda, T, Fukuda, M, Mitsui, Y. | | Deposit date: | 1998-08-06 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure, 7, 1999
|
|
1F88
 
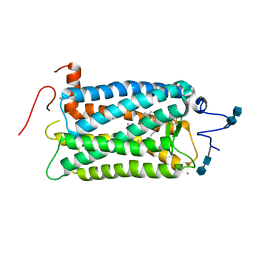 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | Authors: | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
1HCY
 
 | |
1HC1
 
 | |
3VYV
 
 | | Crystal structure of subtilisin NAT at 1.36 | | Descriptor: | CALCIUM ION, GLYCEROL, Subtilisin NAT | | Authors: | Ushijima, H, Fuchita, N, Kajiwara, T, Motoshima, H, Ueno, G, Watanabe, K. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of subtilisin NAT at 1.36
TO BE PUBLISHED
|
|
6AHP
 
 | | Structure of the 58-167 fragment of FliL | | Descriptor: | Flagellar protein FliL | | Authors: | Takekawa, N, Isumi, M, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure ofVibrioFliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function.
MBio, 10, 2019
|
|
