3P96
 
 | |
3ODU
 
 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | 著者 | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | 登録日 | 2010-08-11 | | 公開日 | 2010-10-27 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE6
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | 著者 | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | 登録日 | 2010-08-12 | | 公開日 | 2010-10-27 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | 分子名称: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | 著者 | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | 登録日 | 2010-08-12 | | 公開日 | 2010-10-27 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3PFD
 
 | |
3PGX
 
 | |
3MBM
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with cytosine and FoL fragment 717, imidazo[2,1-b][1,3]thiazol-6-ylmethanol | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | 著者 | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2010-03-25 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3PR0
 
 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptane-1,1-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | 登録日 | 2010-11-29 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3PXU
 
 | |
3PPM
 
 | | Crystal Structure of a Noncovalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | 1-DODECANOL, 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptan-1-one, CHLORIDE ION, ... | | 著者 | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | 登録日 | 2010-11-24 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3A
 
 | |
3PXX
 
 | |
3MD0
 
 | |
2WPD
 
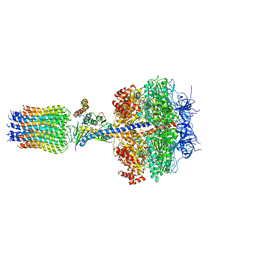 | | The Mg.ADP inhibited state of the yeast F1c10 ATP synthase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE SUBUNIT 9, ... | | 著者 | Dautant, A, Velours, J, Giraud, M.-F. | | 登録日 | 2009-08-05 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.432 Å) | | 主引用文献 | Crystal Structure of the Mg.Adp-Inhibited State of the Yeast F1C10-ATP Synthase.
J.Biol.Chem., 285, 2010
|
|
6TKO
 
 | |
4SBV
 
 | |
2GD1
 
 | |
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | 著者 | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | 登録日 | 1998-04-30 | | 公開日 | 1998-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
8S65
 
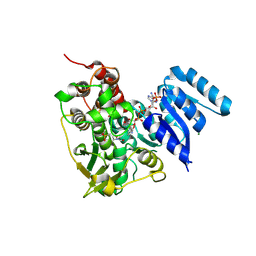 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) as target for anti Toxoplasma gondii compounds: crystal structure, biochemical characterization and biological evaluation of inhibitors | | 分子名称: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CHLORIDE ION, ... | | 著者 | Mazzone, F, Hoeppner, A, Reiners, J, Applegate, V, Abdullaziz, M, Gottstein, J, Wesemann, M, Kurz, T, Smits, S.H, Pfeffer, K. | | 登録日 | 2024-02-26 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-04 | | 実験手法 | SOLUTION SCATTERING (2.56 Å), X-RAY DIFFRACTION | | 主引用文献 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase as target for anti Toxoplasma gondii agents: crystal structure, biochemical characterization and biological evaluation of inhibitors.
Biochem.J., 481, 2024
|
|
1GD1
 
 | |
5A8E
 
 | | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-[(2S)-3-(tert-butylamino)-2-hydroxypropoxy]-7-methyl-1H-indole-2-carbonitrile, ... | | 著者 | Sato, T, Baker, J.G, Warne, T, Brown, G.A, Congreve, M, Leslie, A.G.W, Tate, C.G. | | 登録日 | 2015-07-15 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor.
Mol.Pharmacol., 88, 2015
|
|
2V87
 
 | |
2V83
 
 | |
2V88
 
 | |
2V89
 
 | |
