8GF9
 
 | | Cryo-EM structure of human TRPV1 in cNW11 nanodisc and POPC:POPE:POPG lipids | | 分子名称: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, ... | | 著者 | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-05-10 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
6YKG
 
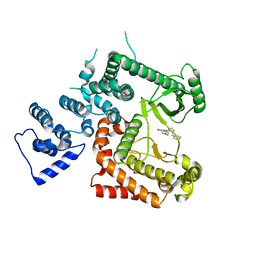 | |
1QMR
 
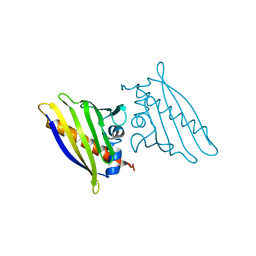 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | 分子名称: | MAJOR POLLEN ALLERGEN BET V 1-A | | 著者 | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | 登録日 | 1999-10-06 | | 公開日 | 2000-10-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
6NPO
 
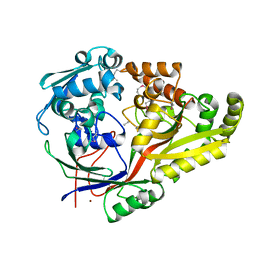 | | Crystal structure of oligopeptide ABC transporter from Bacillus anthracis str. Ames (substrate-binding domain) | | 分子名称: | Oligopeptide ABC transporter, oligopeptide-binding protein, Unknown peptide ligand, ... | | 著者 | Michalska, K, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-01-18 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of oligopeptide ABC transporter from
Bacillus anthracis str. Ames (substrate-binding domain)
To Be Published
|
|
3O1Y
 
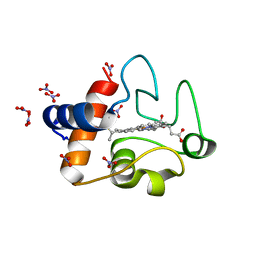 | | Electron transfer complexes: Experimental mapping of the redox-dependent cytochrome c electrostatic surface | | 分子名称: | Cytochrome c, HEME C, NITRATE ION | | 著者 | De March, M, De Zorzi, R, Demitri, N, Gabbiani, C, Guerri, A, Casini, A, Messori, L, Geremia, S. | | 登録日 | 2010-07-22 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
6NAZ
 
 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | 著者 | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | 登録日 | 2018-12-06 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.081 Å) | | 主引用文献 | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | 著者 | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
8GMO
 
 | |
6YM3
 
 | | Crystal structure of Compound 1 with PIP4K2A | | 分子名称: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
5MBG
 
 | | Structure of a bacterial light-regulated adenylyl cyclase | | 分子名称: | Beta subunit of photoactivated adenylyl cyclase, IODIDE ION | | 著者 | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | 登録日 | 2016-11-08 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
6YM4
 
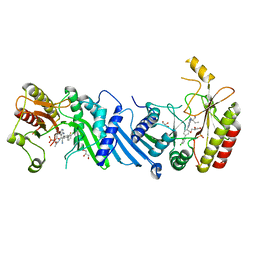 | | Crystal structure of BAY-297 with PIP4K2A | | 分子名称: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | 著者 | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | 登録日 | 2020-04-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
3NRU
 
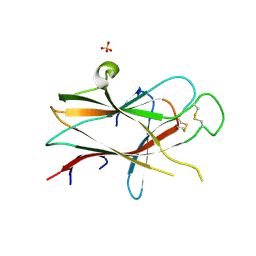 | | Ligand binding domain of EPHA7 | | 分子名称: | CHLORIDE ION, Ephrin receptor, SULFATE ION | | 著者 | Walker, J.R, Yermekbayeva, L, Seitova, A, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | 登録日 | 2010-06-30 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Ephrin A7 ligand binding domain
To be Published
|
|
1Q6D
 
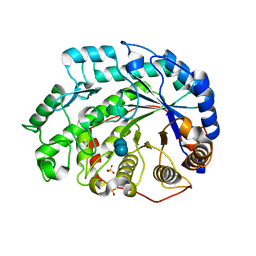 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | 分子名称: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | 登録日 | 2003-08-13 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1L8N
 
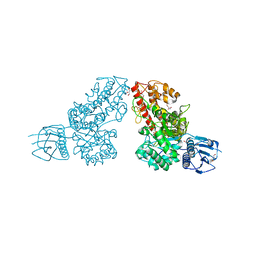 | | The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose | | 分子名称: | 4-O-methyl-beta-D-glucopyranuronic acid, ALPHA-D-GLUCURONIDASE, GLYCEROL, ... | | 著者 | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | 登録日 | 2002-03-21 | | 公開日 | 2003-03-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
6NET
 
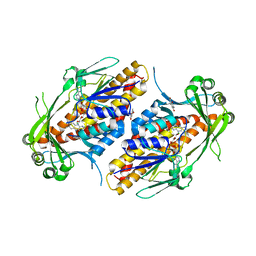 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | 分子名称: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | 著者 | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | 登録日 | 2018-12-18 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NF3
 
 | |
6YPW
 
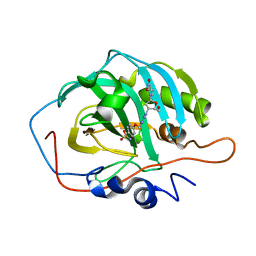 | | Crystal structure for the complex of human carbonic anhydrase II and 4-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide | | 分子名称: | 4-[[1-[(2~{S},3~{S},5~{R})-2-(hydroxymethyl)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]oxolan-3-yl]-1,2,3-triazol-4-yl]methoxy]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Angeli, A, Ferraroni, M. | | 登録日 | 2020-04-16 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
8G2F
 
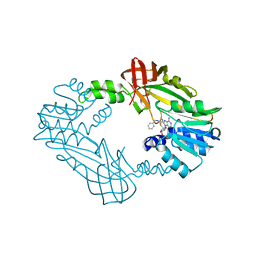 | | Crystal Structure of PRMT3 with Compound II710 | | 分子名称: | 5'-S-[3-(N'-benzylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | 著者 | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-02-03 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
6NDQ
 
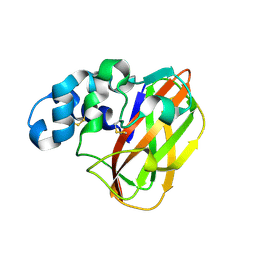 | |
8GFA
 
 | | Cryo-EM structure of human TRPV1 in complex with the analgesic drug SB-366791 | | 分子名称: | (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | 著者 | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-05-10 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
6NIO
 
 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | 著者 | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-12-31 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
6NJX
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with mercury | | 分子名称: | IODIDE ION, MERCURY (II) ION, Xcc_ctr_Hg | | 著者 | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | 登録日 | 2019-01-04 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
8G2W
 
 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-06 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | 分子名称: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | 著者 | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | 登録日 | 2002-05-21 | | 公開日 | 2002-07-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1KYT
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | 分子名称: | CALCIUM ION, hypothetical protein TA0175 | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-02-05 | | 公開日 | 2003-01-21 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
