8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2022-12-14 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4U
 
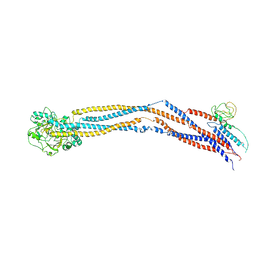 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | 分子名称: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | 著者 | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
5GH9
 
 | |
4OU4
 
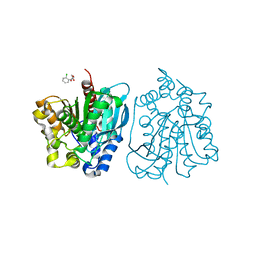 | | Crystal structure of esterase rPPE mutant S159A complexed with (S)-Ac-CPA | | 分子名称: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB6
 
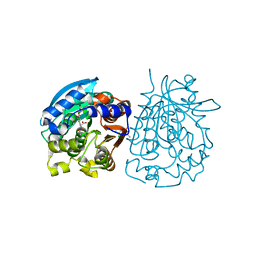 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | 分子名称: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | 分子名称: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | 登録日 | 2022-07-13 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
4OB7
 
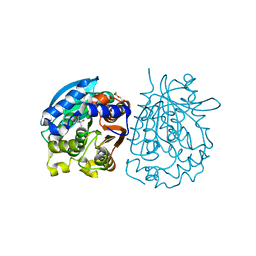 | | Crystal structure of esterase rPPE mutant W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
2JZ3
 
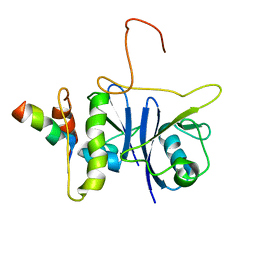 | | SOCS box elonginBC ternary complex | | 分子名称: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | 著者 | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | 登録日 | 2007-12-27 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
4ORD
 
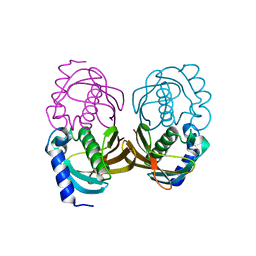 | |
4OU5
 
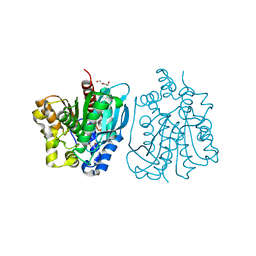 | | Crystal structure of esterase rPPE mutant S159A/W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
2K8H
 
 | |
6G3E
 
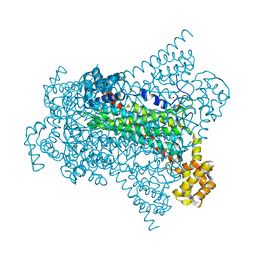 | | Crystal structure of EDDS lyase in complex with formate | | 分子名称: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3D
 
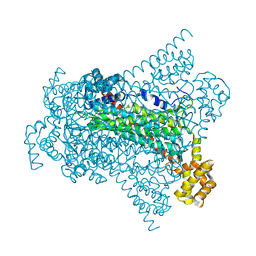 | |
6G3F
 
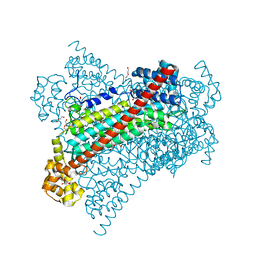 | | Crystal structure of EDDS lyase in complex with fumarate | | 分子名称: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, FUMARIC ACID | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.222 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3I
 
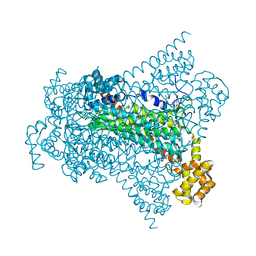 | | Crystal structure of EDDS lyase in complex with N-(2-aminoethyl)aspartic acid (AEAA) | | 分子名称: | (2~{S})-2-(2-azanylethylamino)butanedioic acid, Argininosuccinate lyase, FUMARIC ACID | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3H
 
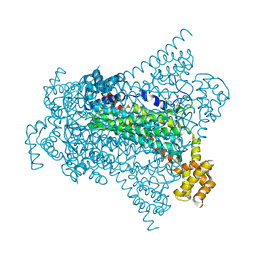 | | Crystal structure of EDDS lyase in complex with SS-EDDS | | 分子名称: | (2~{S})-2-[2-[[(2~{S})-1,4-bis(oxidanyl)-1,4-bis(oxidanylidene)butan-2-yl]amino]ethylamino]butanedioic acid, Argininosuccinate lyase | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.269 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
8IX0
 
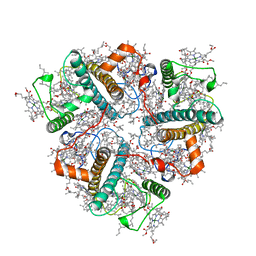 | | Cryo-EM structure of unprotonated LHCII nanodisc at high pH value | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Ruan, M.X, Ding, W. | | 登録日 | 2023-03-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX2
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at low pH value | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Ruan, M.X, Ding, W. | | 登録日 | 2023-03-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
5ED3
 
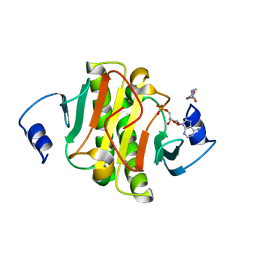 | | crystal structure of human Hint1 complexing with AP5A | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | 著者 | Wang, J, Fang, P, Guo, M. | | 登録日 | 2015-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.309 Å) | | 主引用文献 | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8IWX
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Ruan, M.X, Ding, W. | | 登録日 | 2023-03-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWY
 
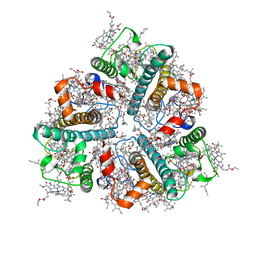 | | Cryo-EM structure of protonated LHCII in detergent solution at low pH value | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Ruan, M.X, Ding, W. | | 登録日 | 2023-03-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
