6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
1HTO
 
 | |
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
1HPT
 
 | |
6ZFM
 
 | | Structure of alpha-Cobratoxin with a peptide inhibitor | | 分子名称: | 3-[2-[2-[2-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propan-1-ol, Alpha-cobratoxin, PENTAETHYLENE GLYCOL, ... | | 著者 | Kiontke, S, Kummel, D. | | 登録日 | 2020-06-17 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Peptide Inhibitors of the alpha-Cobratoxin-Nicotinic Acetylcholine Receptor Interaction.
J.Med.Chem., 63, 2020
|
|
7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6ZIR
 
 | | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP AND COMPOUND 18 | | 分子名称: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kessler, D, Fischer, G, Boettcher, J. | | 登録日 | 2020-06-26 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6ZIW
 
 | | The IRAK3 Pseudokinase Domain Bound To ATPgammaS | | 分子名称: | Interleukin-1 receptor-associated kinase 3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | 著者 | Mathea, S, Chatterjee, D, Preuss, F, Kraemer, A, Knapp, S. | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | The IRAK3 Pseudokinase Domain Bound To ATPgammaS
To Be Published
|
|
6ZJ0
 
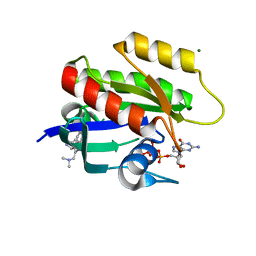 | | CRYSTAL STRUCTURE OF HRAS-G12D IN COMPLEX WITH GCP AND COMPOUND 18 | | 分子名称: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Kessler, D, Fischer, G, Boettcher, J. | | 登録日 | 2020-06-26 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
1HIB
 
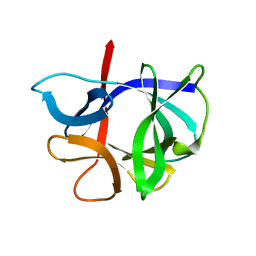 | | THE STRUCTURE OF AN INTERLEUKIN-1 BETA MUTANT WITH REDUCED BIOACTIVITY SHOWS MULTIPLE SUBTLE CHANGES IN CONFORMATION THAT AFFECT PROTEIN-PROTEIN RECOGNITION | | 分子名称: | INTERLEUKIN-1 BETA | | 著者 | Camacho, N.P, Smith, D.R, Goldman, A, Schneider, B, Green, D, Young, P.R, Berman, H.M. | | 登録日 | 1993-03-29 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of an interleukin-1 beta mutant with reduced bioactivity shows multiple subtle changes in conformation that affect protein-protein recognition.
Biochemistry, 32, 1993
|
|
6ZK9
 
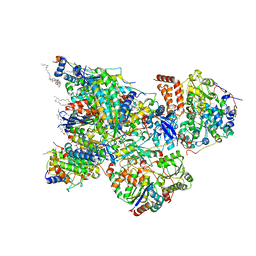 | | Peripheral domain of open complex I during turnover | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKN
 
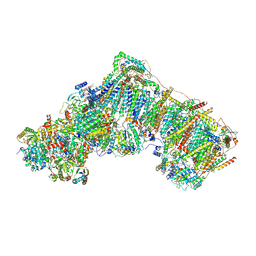 | | Complex I inhibited by rotenone, open3 | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKQ
 
 | | Native complex I, open2 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZI8
 
 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | 分子名称: | CHLORIDE ION, Insulin, ZINC ION | | 著者 | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | 登録日 | 2020-06-25 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZKC
 
 | | Complex I during turnover, closed | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
1VSR
 
 | | VERY SHORT PATCH REPAIR (VSR) ENDONUCLEASE FROM ESCHERICHIA COLI | | 分子名称: | PROTEIN (VSR ENDONUCLEASE), ZINC ION | | 著者 | Tsutakawa, S.E, Muto, T, Jingami, H, Kunishima, N, Ariyoshi, M, Kohda, D, Nakagawa, M, Morikawa, K. | | 登録日 | 1999-02-13 | | 公開日 | 1999-10-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic and functional studies of very short patch repair endonuclease.
Mol.Cell, 3, 1999
|
|
6ZKR
 
 | | Native complex I, open3 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6YT6
 
 | |
1VT8
 
 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | 分子名称: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | 著者 | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | 登録日 | 1996-12-02 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
1HKO
 
 | |
6YXV
 
 | |
6YVY
 
 | |
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | 分子名称: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | 著者 | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | 登録日 | 2001-01-22 | | 公開日 | 2001-04-04 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
