8JIL
 
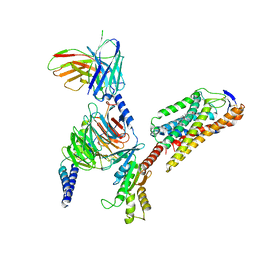 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JII
 
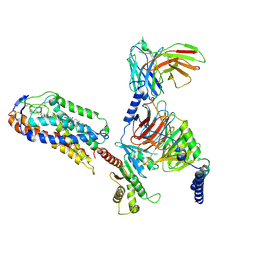 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JHY
 
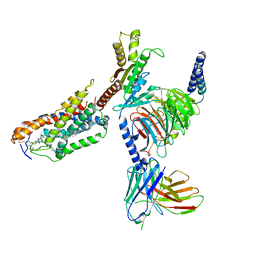 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-25 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
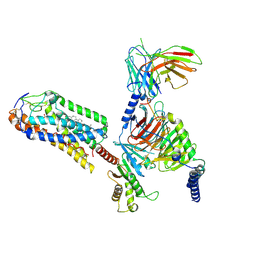 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
2GM1
 
 | | Crystal structure of the mitotic kinesin eg5 in complex with mg-adp and n-(3-aminopropyl)-n-((3-benzyl-5-chloro-4-oxo-3,4-dihydropyrrolo[2,1-f][1,2,4]triazin-2-yl)(cyclopropyl)methyl)-4-methylbenzamide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-RELATED MOTOR PROTEIN EG5, MAGNESIUM ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2006-04-05 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis and SAR of pyrrolotriazine-4-one based Eg5 inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | 著者 | Ting, Y.T, Broury, A, Ooi, J. | | 登録日 | 2023-06-29 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.12621117 Å) | | 主引用文献 | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
8JYS
 
 | |
8WMI
 
 | |
8WMC
 
 | |
8WM4
 
 | |
8WML
 
 | |
8P1U
 
 | | Structure of divisome complex FtsWIQLB | | 分子名称: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | 著者 | Yang, L, Chang, S, Tang, D, Dong, H. | | 登録日 | 2023-05-12 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
6BFN
 
 | | Crystal structure of human IRAK1 | | 分子名称: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | 著者 | Wang, L, Qiao, Q, Wu, H. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | 著者 | Wu, Y, Qi, J, Gao, F. | | 登録日 | 2020-04-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | 分子名称: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | 著者 | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | 登録日 | 2020-07-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7C7P
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | 分子名称: | 3C-like proteinase, boceprevir (bound form) | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | 登録日 | 2020-08-04 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D5K
 
 | | CryoEM structure of cotton cellulose synthase isoform 7 | | 分子名称: | Cellulose synthase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Guan, Z.Y, Xue, Y, Yin, P, Zhang, X.L. | | 登録日 | 2020-09-26 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum.
Plant Biotechnol J, 19, 2021
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | 著者 | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | 登録日 | 2020-09-19 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Wang, X, Ma, L, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K8I
 
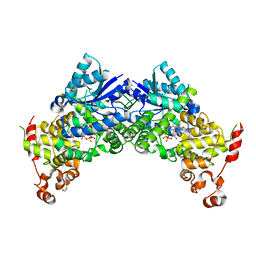 | | Crystal structure of Arabidopsis thaliana CRY2 | | 分子名称: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Ma, L, Wang, X, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7CRU
 
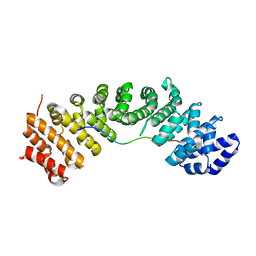 | | hnRNPK NLS in complex with Importin alpha 1 (KPNA2) | | 分子名称: | ACETATE ION, GLYCEROL, Heterogeneous nuclear ribonucleoprotein K, ... | | 著者 | Yao, J, Sun, Q. | | 登録日 | 2020-08-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nuclear import receptors and hnRNPK mediates nuclear import and stress granule localization of SIRLOIN.
Cell.Mol.Life Sci., 78, 2021
|
|
7CRE
 
 | |
