5EBX
 
 | |
3EBX
 
 | | REFINEMENT AT 1.4 ANGSTROMS RESOLUTION OF A MODEL OF ERABUTOXIN B. TREATMENT OF ORDERED SOLVENT AND DISCRETE DISORDER | | 分子名称: | ERABUTOXIN B, SULFATE ION | | 著者 | Smith, J.L, Corfield, P.W.R, Hendrickson, W.A, Low, B.W. | | 登録日 | 1988-01-15 | | 公開日 | 1988-04-16 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
5X3D
 
 | | Crystal structure of HEP-CMP-bound form of cytidylyltransferase (CyTase) domain of Fom1 from Streptomyces wedmorensis | | 分子名称: | Phosphoenolpyruvate phosphomutase, [[(2R,3S,4R,5R)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-(2-hydroxyethyl)phosphinic acid | | 著者 | Tomita, T, Cho, S.H, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2017-02-04 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Fosfomycin Biosynthesis via Transient Cytidylylation of 2-Hydroxyethylphosphonate by the Bifunctional Fom1 Enzyme
ACS Chem. Biol., 12, 2017
|
|
5XH8
 
 | |
5XH9
 
 | | Aspergillus kawachii beta-fructofuranosidase | | 分子名称: | Extracellular invertase, SODIUM ION | | 著者 | Nagaya, M, Tonozuka, T. | | 登録日 | 2017-04-19 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Biosci. Biotechnol. Biochem., 81, 2017
|
|
2RR6
 
 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | 分子名称: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | 著者 | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | 登録日 | 2010-05-25 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
8HAL
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 1 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
3SSI
 
 | |
8J91
 
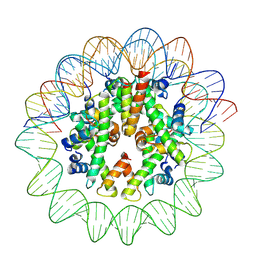 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | 分子名称: | DNA (169-MER), HTA13, Histone H2B.6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
8J90
 
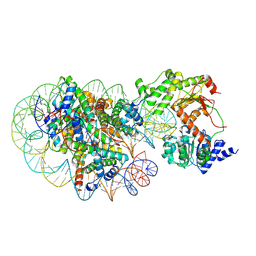 | | Cryo-EM structure of DDM1-nucleosome complex | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (169-MER), HTA6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.71 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
8J92
 
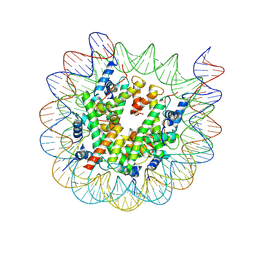 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W | | 分子名称: | DNA (169-MER), HTA6, HTB9, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
6LUD
 
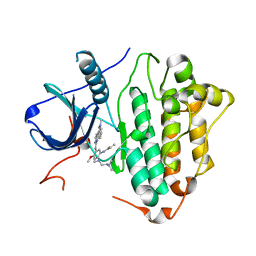 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with Osimertinib | | 分子名称: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | 著者 | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | 登録日 | 2020-01-27 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
4E1B
 
 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | 分子名称: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | 著者 | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | 登録日 | 2012-03-06 | | 公開日 | 2012-03-14 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6LUB
 
 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 | | 分子名称: | Epidermal growth factor receptor, N-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-7-(4-methylpiperazin-1-yl)-5-propan-2-yl-9-[2,2,2-tris(fluoranyl)ethoxy]pyrido[4,3-b]indol-3-amine | | 著者 | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | 登録日 | 2020-01-27 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.315 Å) | | 主引用文献 | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
8I5L
 
 | |
8JG5
 
 | | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | | 分子名称: | VH,SARAH, VL,SARAH, VP1 | | 著者 | Hosaka, T, Katsura, K, Kimura-Someya, T, Someya, Y, Shirouzu, M. | | 登録日 | 2023-05-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography revealing binding sites for human monoclonal antibodies.
J.Virol., 98, 2024
|
|
1M3A
 
 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | 分子名称: | Proto-oncogene C-crk | | 著者 | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | 登録日 | 2002-06-27 | | 公開日 | 2003-08-05 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | 分子名称: | Proto-oncogene C-crk | | 著者 | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | 登録日 | 2002-06-26 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
