7BD9
 
 | | Notum Fragment 886 | | 分子名称: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-21 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8Y
 
 | | Notum-Fragment 276 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(3-ethoxyphenyl)-1,3,4-thiadiazol-2-amine, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jonees, E.Y. | | 登録日 | 2020-12-13 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9D
 
 | | Notum Fragment 290 | | 分子名称: | 2-(4-acetamidophenoxy)ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-14 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDC
 
 | | Notum Fragment 923 | | 分子名称: | 1-methyl-N-[(2-methylphenyl)methyl]-1H-tetrazol-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-21 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7COF
 
 | |
7COG
 
 | |
4EF3
 
 | | Multicopper Oxidase CueO (Citrate buffer) | | 分子名称: | Blue copper oxidase CueO, CITRIC ACID, COPPER (II) ION, ... | | 著者 | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | 登録日 | 2012-03-29 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Exogenous acetate ion reaches the type II copper centre in CueO through the water-excretion channel and potentially affects the enzymatic activity.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6MBZ
 
 | | Structure of Transcription Factor | | 分子名称: | Signal transducer and activator of transcription 5B | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2018-08-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural and functional consequences of the STAT5BN642Hdriver mutation.
Nat Commun, 10, 2019
|
|
6MBW
 
 | | Structure of Transcription Factor | | 分子名称: | Signal transducer and activator of transcription 5B | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2018-08-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structural and functional consequences of the STAT5BN642H driver mutation.
Nat Commun, 10, 2019
|
|
7B99
 
 | | Notum Fragment 283 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-14 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8U
 
 | | Notum-Fragment 201 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(piperidin-1-yl)-1,2,5-oxadiazol-3-amine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Zhao, Y, Jonees, E.Y. | | 登録日 | 2020-12-13 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8M
 
 | | Notum-Fragment 193 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methoxy-2-(1~{H}-pyrazol-3-yl)phenol, ... | | 著者 | Zhao, Y, Jonees, E.Y. | | 登録日 | 2020-12-13 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9I
 
 | | Notum Fragment 297 | | 分子名称: | 1-(1,3-benzodioxol-5-yl)-~{N}-(pyridin-2-ylmethyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-14 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BA1
 
 | | Notum Fragment 634 | | 分子名称: | 1-ethanoylpiperidine-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-15 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCD
 
 | | Notum Fragment 714 | | 分子名称: | 2-(morpholin-4-ium-4-ylmethyl)naphthalen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-19 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9N
 
 | | Notum Fragment 588 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2020-12-14 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
3CG7
 
 | |
3CM6
 
 | |
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | 著者 | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | 登録日 | 1992-01-15 | | 公開日 | 1992-07-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
6JXD
 
 | |
6K1J
 
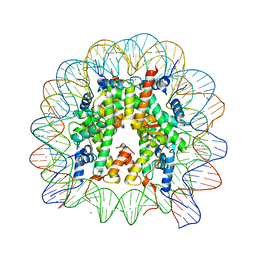 | | Human nucleosome core particle with H2A.X variant | | 分子名称: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | 著者 | Sharma, D, De Falco, L, Davey, C.A. | | 登録日 | 2019-05-10 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
6LCS
 
 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | 分子名称: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | 著者 | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | 登録日 | 2019-11-19 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
6K1K
 
 | | Human nucleosome core particle with H2A.X S139E variant | | 分子名称: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | 著者 | Sharma, D, De Falco, L, Davey, C.A. | | 登録日 | 2019-05-10 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
6K1I
 
 | | Human nucleosome core particle with gammaH2A.X variant | | 分子名称: | CHLORIDE ION, DNA (147-MER), Histone H2AX, ... | | 著者 | Sharma, D, De Falco, L, Davey, C.A. | | 登録日 | 2019-05-10 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
2GOV
 
 | | Solution structure of Murine p22HBP | | 分子名称: | Heme-binding protein 1 | | 著者 | Volkman, B.F, Dias, J.S, Goodfellow, B.J, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-14 | | 公開日 | 2006-05-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
J.Biol.Chem., 281, 2006
|
|
