6RYB
 
 | |
3LQR
 
 | | Structure of CED-4:CED-3 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.896 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
5D39
 
 | | Transcription factor-DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-08-06 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6RYA
 
 | |
6FBV
 
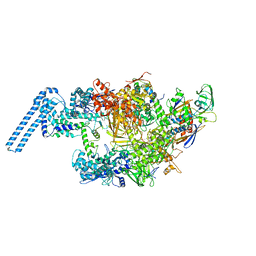 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Das, K, Lin, W, Ebright, E. | | 登録日 | 2017-12-19 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
5OF9
 
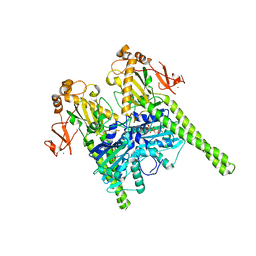 | | Crystal structure of human MORC2 (residues 1-603) | | 分子名称: | MAGNESIUM ION, MORC family CW-type zinc finger protein 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Douse, C.H, Shamin, M, Modis, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.807 Å) | | 主引用文献 | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
3PUQ
 
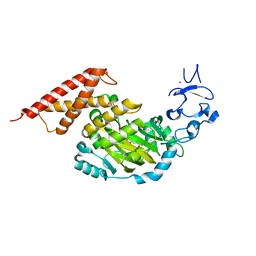 | | CEKDM7A from C.Elegans, complex with alpha-KG | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
3PUR
 
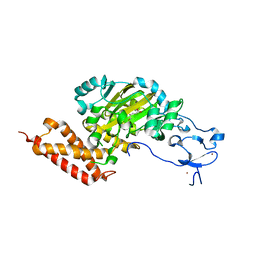 | | CEKDM7A from C.Elegans, complex with D-2-HG | | 分子名称: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
4Y5U
 
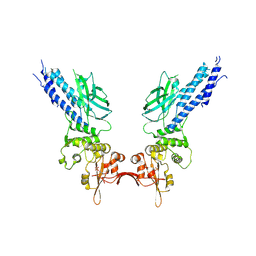 | | Transcription factor | | 分子名称: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7D14
 
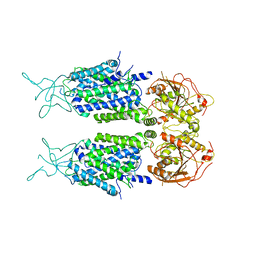 | | Mouse KCC2 | | 分子名称: | Solute carrier family 12 member 5 | | 著者 | Zhang, S, Yang, M. | | 登録日 | 2020-09-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
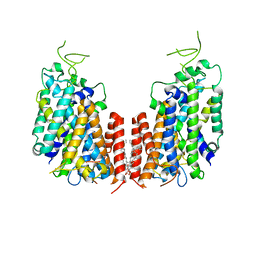 | | Human NKCC1 | | 分子名称: | PALMITIC ACID, Solute carrier family 12 member 2 | | 著者 | Zhang, S, Yang, M. | | 登録日 | 2020-09-12 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7CYB
 
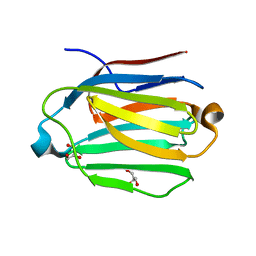 | |
7CYA
 
 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | 分子名称: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J. | | 登録日 | 2020-09-03 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
5X80
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 COMPLEX WITH SALICYLIC ACID | | 分子名称: | 2-HYDROXYBENZOIC ACID, SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | 著者 | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | 登録日 | 2017-02-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5X88
 
 | | A crystal structure of cutinases from Malbranchea cinnamomea | | 分子名称: | cutinase | | 著者 | Jiang, Z, Yang, S.Q, You, X, Huang, P, Ma, J.W. | | 登録日 | 2017-03-01 | | 公開日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | High-level expression and characterization of a novel cutinase from Malbranchea cinnamomea suitable for butyl butyrate production.
Biotechnol Biofuels, 10, 2017
|
|
7C7P
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | 分子名称: | 3C-like proteinase, boceprevir (bound form) | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | 登録日 | 2020-08-04 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
4UZ7
 
 | |
4UZ5
 
 | |
4WCM
 
 | |
4UZ6
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | 分子名称: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Zebisch, M, Jones, E.Y. | | 登録日 | 2014-09-04 | | 公開日 | 2015-02-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4UZQ
 
 | |
4WCL
 
 | |
4WCN
 
 | | Crystal Structure of Tripeptide bound Cell Shape Determinant Csd4 protein from Helicobacter pylori | | 分子名称: | Conserved hypothetical secreted protein, IODIDE ION, N-acetyl-L-alanyl-N-[(1S,5R)-5-amino-1,5-dicarboxypentyl]-D-glutamine, ... | | 著者 | Chan, A.C, Murphy, M.E. | | 登録日 | 2014-09-05 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Helical Shape of Helicobacter pylori Requires an Atypical Glutamine as a Zinc Ligand in the Carboxypeptidase Csd4.
J.Biol.Chem., 290, 2015
|
|
