5O5X
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | 分子名称: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | 著者 | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | 登録日 | 2017-06-02 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.148 Å) | | 主引用文献 | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Y
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | 分子名称: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | 著者 | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | 登録日 | 2017-06-02 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.915 Å) | | 主引用文献 | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7U8F
 
 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | 分子名称: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | 著者 | Ma, X, Ornelas, E, Clifton, M.C. | | 登録日 | 2022-03-08 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
6LXY
 
 | | IRAK4 in complex with inhibitor | | 分子名称: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-fluoranyl-3-methyl-3-oxidanyl-butyl]-6-[(6-fluoranylpyrazolo[1,5-a]pyrimidin-5-yl)amino]-4-(propan-2-ylamino)pyridine-3-carboxamide, SULFATE ION | | 著者 | Ghosh, K, Bose, S. | | 登録日 | 2020-02-12 | | 公開日 | 2020-11-25 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis.
Acs Med.Chem.Lett., 11, 2020
|
|
1EGO
 
 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | 分子名称: | GLUTAREDOXIN | | 著者 | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | 登録日 | 1991-10-08 | | 公開日 | 1993-10-31 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1ESP
 
 | | NEUTRAL PROTEASE MUTANT E144S | | 分子名称: | CALCIUM ION, NEUTRAL PROTEASE MUTANT E144S, ZINC ION | | 著者 | Litster, S.A, Wetmore, D.R, Roche, R.S, Codding, P.W. | | 登録日 | 1995-08-11 | | 公開日 | 1995-12-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | E144S active-site mutant of the Bacillus cereus thermolysin-like neutral protease at 2.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1ED4
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH IPITU (H4B FREE) | | 分子名称: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | 登録日 | 2000-01-26 | | 公開日 | 2000-10-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
1ED5
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH NNA(H4B FREE) | | 分子名称: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | 登録日 | 2000-01-26 | | 公開日 | 2001-01-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | 分子名称: | GLUTAREDOXIN | | 著者 | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | 登録日 | 1991-10-08 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
8FNR
 
 | |
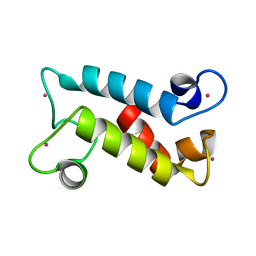
8FNS
 
 | |
3LYZ
 
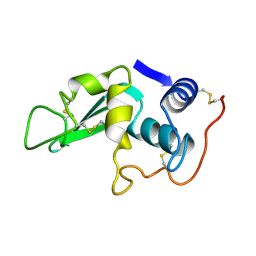 | |
7KDT
 
 | |
7OAO
 
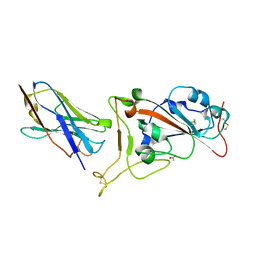 | | Nanobody C5 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
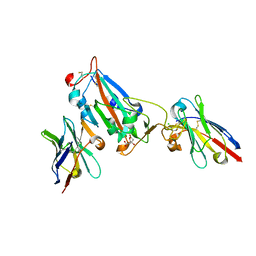 | |
7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
 | | Nanobody C5 bound to Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Naismith, J.H, Weckener, M. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-20 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | |
7XCL
 
 | |
7XCM
 
 | |
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | 分子名称: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | 著者 | Li, J, Chan, M.K. | | 登録日 | 2022-03-24 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
5HJX
 
 | | Structure function studies of R. palustris RubisCO (A47V mutant; CABP-bound) | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | 著者 | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | 登録日 | 2016-01-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HQL
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant; CABP-bound; no expression tag) | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | 著者 | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | 登録日 | 2016-01-21 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structure function studies of R. palustris RubisCO
To Be Published
|
|
6W7F
 
 | | Structure of EED bound to inhibitor 5285 | | 分子名称: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | 著者 | Petrunak, E.M, Stuckey, J.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
